LGCPs - Distance sampling
David Borchers and Finn Lindgren
Generated on 2026-02-26
Source:vignettes/articles/2d_lgcp_distancesampling.Rmd
2d_lgcp_distancesampling.RmdIntroduction
We’re going to estimate distribution and abundance from a line
transect survey of dolphins in the Gulf of Mexico. These data are also
available in the R package dsm (where they go
under the name mexdolphins). In inlabru the
data are called mexdolphin for sp format, and
mexdolphin_sf for sf format.
Get the data
We’ll start by loading the data, renaming it, and extracting the mesh (for convenience).
mexdolphin <- mexdolphin_sf
mesh <- mexdolphin$meshPlot the data (the initial code below is just to get rid of tick marks, if desired)
noyticks <- theme(
axis.text.y = element_blank(),
axis.ticks = element_blank()
)
noxticks <- theme(
axis.text.x = element_blank(),
axis.ticks = element_blank()
)
ggplot() +
gg(mexdolphin$ppoly) +
gg(mexdolphin$samplers, color = "grey") +
gg(mexdolphin$points, size = 0.2, alpha = 1) +
theme(
legend.key.width = unit(x = 0.2, "cm"),
legend.key.height = unit(x = 0.3, "cm")
) +
theme(legend.text = element_text(size = 6)) +
coord_sf(datum = fm_crs(mexdolphin$ppoly))
Spatial model with a half-normal detection function
The samplers in this dataset are lines, not polygons, so
we need to tell inlabru about the strip half-width,
W, which in the case of these data is 8. We start by
plotting the distances and histogram of frequencies in distance
intervals:
W <- 8
ggplot(mexdolphin$points) +
geom_histogram(aes(x = distance),
breaks = seq(0, W, length.out = 9),
boundary = 0, fill = NA, color = "black"
) +
geom_point(aes(x = distance), y = 0, pch = "|", cex = 4)
We need to define a half-normal detection probability function. This must take distance as its first argument and the linear predictor of the sigma parameter as its second:
hn <- function(distance, sigma) {
exp(-0.5 * (distance / sigma)^2)
}To control the prior distribution for the sigma
parameter, we use a transformation mapper that converts a N(0, 1) latent variable into an exponentially
distributed variable with expectation 8 (this is a somewhat arbitrary
value, but motivated by the maximum observation distance
W):
bm_marginal(qexp, pexp, dexp, rate = 1 / 8)Specify and fit an SPDE model to these data using a half-normal detection function form. We need to define a (Matérn) covariance function for the SPDE:
matern <- inla.spde2.pcmatern(mexdolphin$mesh,
prior.sigma = c(2, 0.01),
prior.range = c(50, 0.01)
)Here, the range is probabilistically limited by P(\text{range}\leq 50)=0.01 and the standard deviation of the spatial field is limited by P(\text{sd}\geq 2)=0.01.
We need to now separately define the components of the model (the
SPDE, the Intercept and the detection function parameter
sigma)
cmp <- ~ mySPDE(main = geometry, model = matern) +
sigma(1,
prec.linear = 1,
marginal = bm_marginal(qexp, pexp, dexp, rate = 1 / 8)
) +
Intercept(1)The marginal argument in the sigma
component specifies the transformation function taking N(0,1) to
Exponential(1/8).
The formula, which describes how these components are combined to form the linear predictor (remembering that we need an offset due to the unknown direction of the detections!):
Before version 2.9.0.9004, a less compact approach had
to be used, by applying the transformation between N(0,1) and
Exponential(1/8) directly in the predictor expression, requiring
corresponding adjustments to the later predict() calls,
etc:
# sigma_transf <- function(x) {
# bru_forward_transformation(qexp, x, rate = 1 / 8)
# }
# cmp <- ~ mySPDE(main = geometry, model = matern) +
# sigma_theta(1, prec.linear = 1) +
# Intercept(1)
# form <- geometry + distance ~ mySPDE +
# log(hn(distance, sigma_transf(sigma_theta))) +
# Intercept + log(2)Then we fit the model, passing both the components and the formula
(previously the formula was constructed invisibly by
inlabru), and specify integration domains for the spatial
and distance dimensions:
fit <- lgcp(
components = cmp,
mexdolphin$points,
samplers = mexdolphin$samplers,
domain = list(
geometry = mesh,
distance = fm_mesh_1d(seq(0, 8, length.out = 30))
),
formula = form
)Look at the SPDE parameter posteriors
spde.range <- spde.posterior(fit, "mySPDE", what = "range")
plot(spde.range)
spde.logvar <- spde.posterior(fit, "mySPDE", what = "log.variance")
plot(spde.logvar)
Predict spatial intensity, and plot it:
pxl <- fm_pixels(mesh, dims = c(200, 100), mask = mexdolphin$ppoly)
pr.int <- predict(fit, pxl, ~ exp(mySPDE + Intercept))
ggplot() +
gg(pr.int, geom = "tile") +
gg(mexdolphin$ppoly, linewidth = 1, alpha = 0) +
gg(mexdolphin$samplers, color = "grey") +
gg(mexdolphin$points, size = 0.2, alpha = 1) +
theme(
legend.key.width = unit(x = 0.2, "cm"),
legend.key.height = unit(x = 0.3, "cm")
) +
theme(legend.text = element_text(size = 6))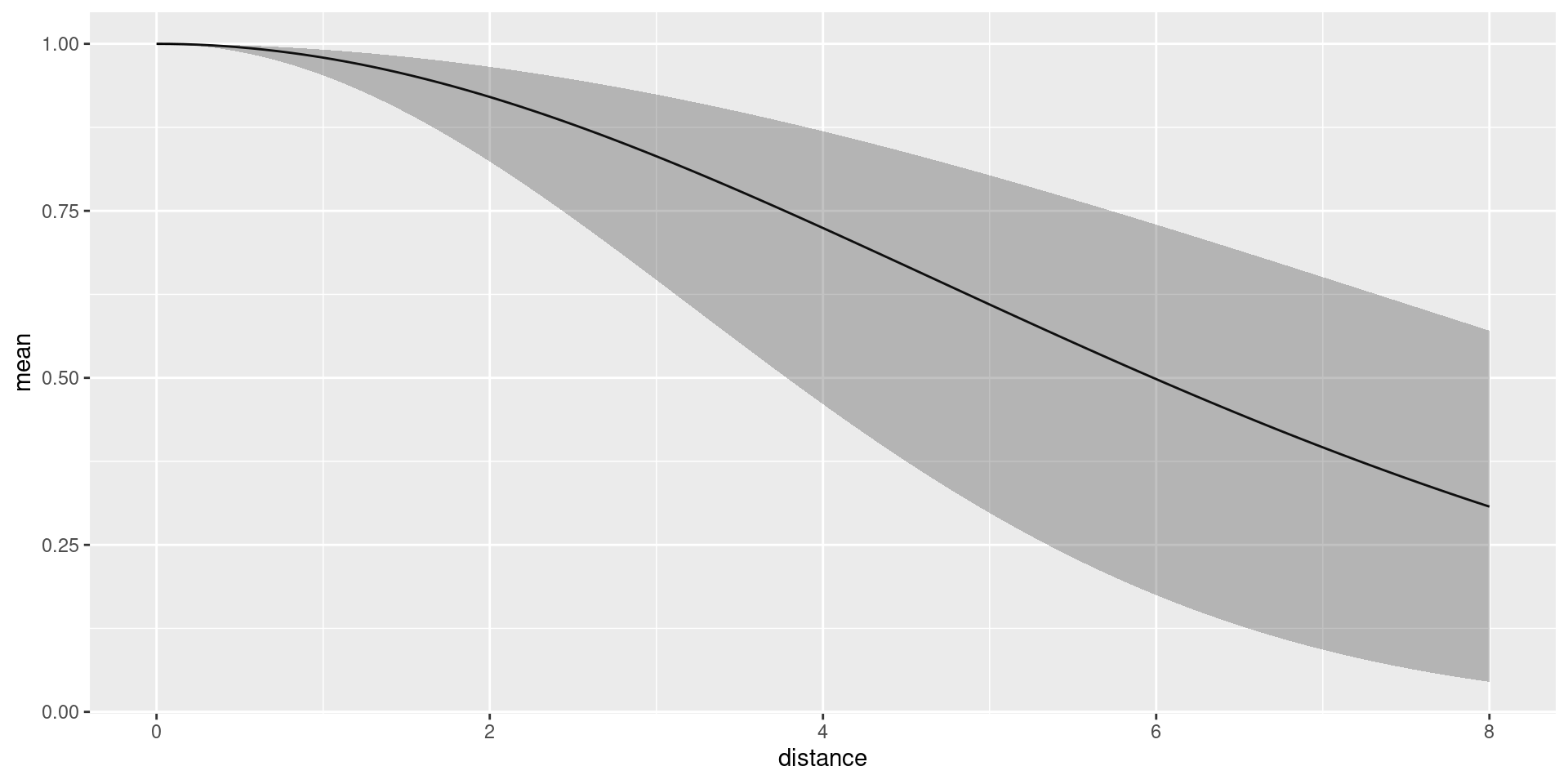
Predict the detection function and plot it, to generate a plot like
the one below. Here, we should make sure that it doesn’t try to evaluate
the effects of components that can’t be evaluated using the given input
data. From version 2.8.0, inlabru automatically detects
which components are involved. See ?predict.bru for more
information.
distdf <- data.frame(distance = seq(0, 8, length.out = 100))
dfun <- predict(fit, distdf, ~ hn(distance, sigma))
plot(dfun)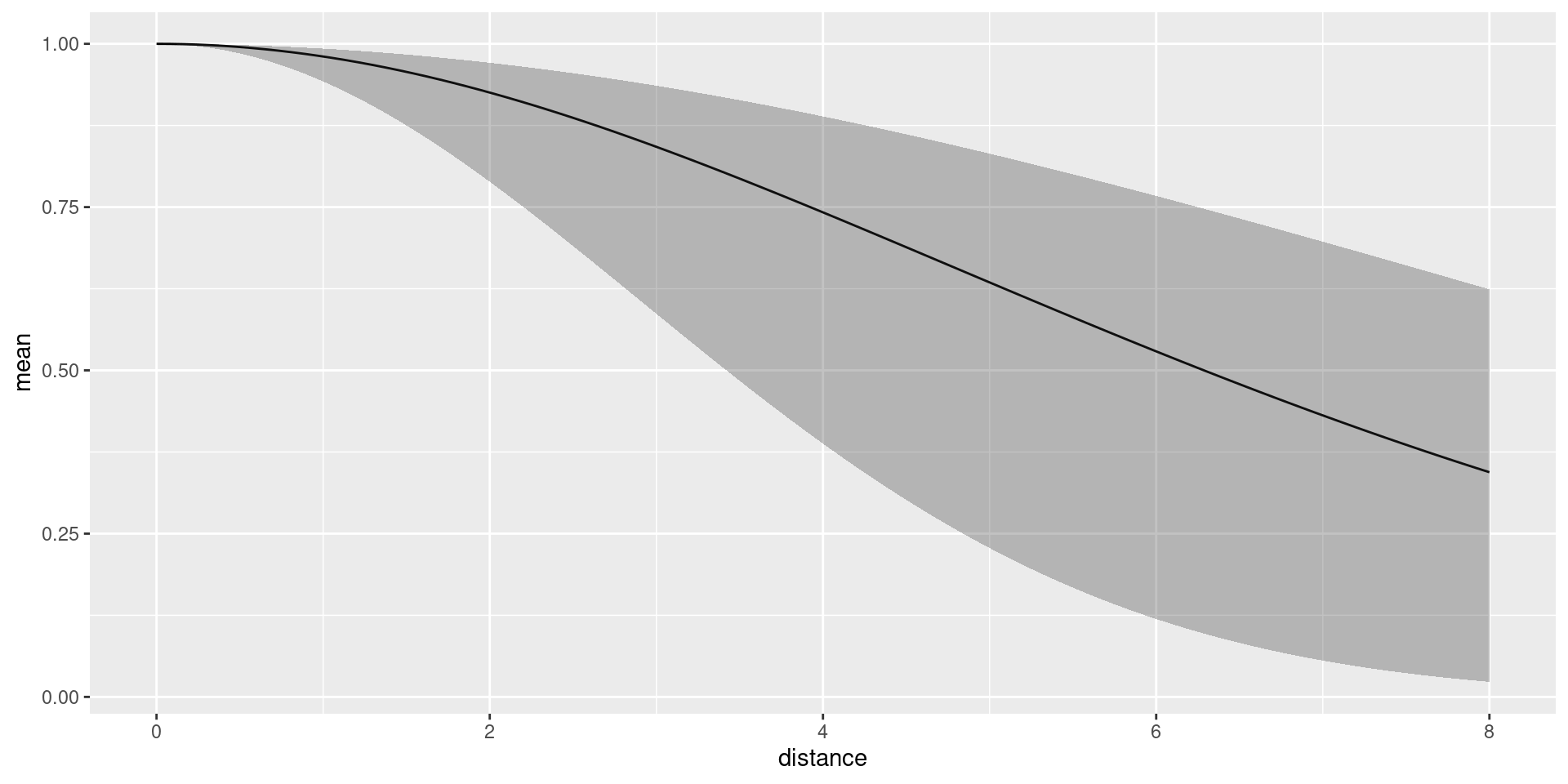 The average detection probability within the maximum detection distance
is estimated to be 0.7136295.
The average detection probability within the maximum detection distance
is estimated to be 0.7136295.
We can look at the posterior for expected number of dolphins as usual:
predpts <- fm_int(mexdolphin$mesh, mexdolphin$ppoly)
Lambda <- predict(fit, predpts, ~ sum(weight * exp(mySPDE + Intercept)))
Lambda
#> mean sd q0.025 q0.5 q0.975 median mean.mc_std_err
#> 1 248.4814 52.17527 163.4862 245.4218 350.5155 245.4218 5.951824
#> sd.mc_std_err
#> 1 3.671481and including the randomness about the expected number. In this case, it turns out that you need lots of posterior samples, e.g. 2,000 to smooth out the Monte Carlo error in the posterior, and this takes a little while to compute:
Ns <- seq(50, 450, by = 1)
Nest <- predict(fit, predpts,
~ data.frame(
N = Ns,
density = dpois(
Ns,
lambda = sum(weight * exp(mySPDE + Intercept))
)
),
n.samples = 2000
)
Nest <- dplyr::bind_rows(
cbind(Nest, Method = "Posterior"),
data.frame(
N = Nest$N,
mean = dpois(Nest$N, lambda = Lambda$mean),
mean.mc_std_err = 0,
Method = "Plugin"
)
)
ggplot(data = Nest) +
geom_line(aes(x = N, y = mean, colour = Method)) +
geom_ribbon(
aes(
x = N,
ymin = mean - 2 * mean.mc_std_err,
ymax = mean + 2 * mean.mc_std_err,
fill = Method,
),
alpha = 0.2
) +
geom_line(aes(x = N, y = mean, colour = Method)) +
ylab("Probability mass function")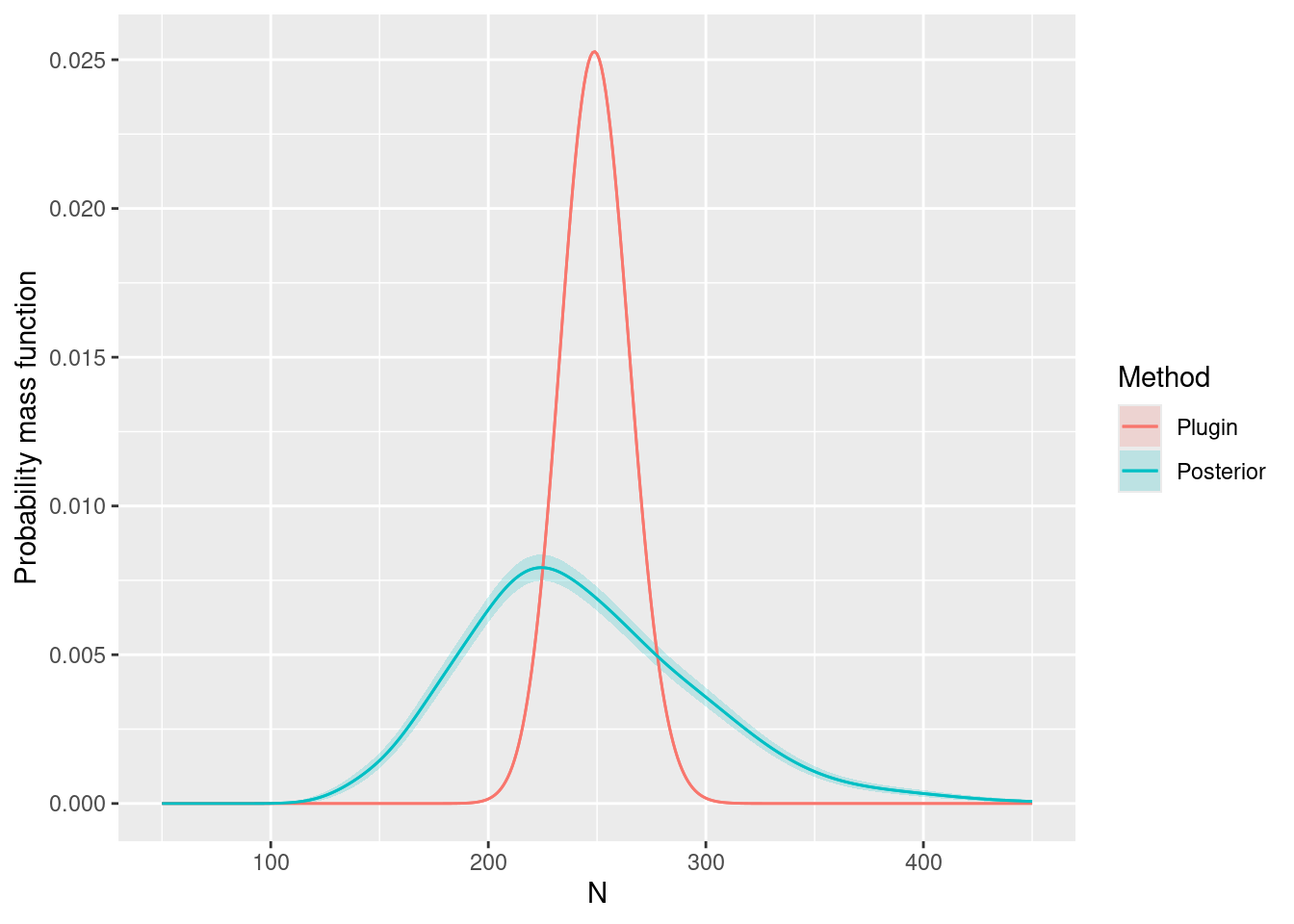
Hazard-rate Detection Function
Try doing this all again, but use this hazard-rate detection function
model, with the same prior for the sigma parameter as for
the half-Normal model (such parameters aren’t always comparable, but in
this example it’s a reasonable choice):
hr <- function(distance, sigma) {
1 - exp(-(distance / sigma)^-1)
}Solution:
formula1 <- geometry + distance ~ mySPDE +
log(hr(distance, sigma)) +
Intercept + log(2)
fit1 <- lgcp(
components = cmp,
mexdolphin$points,
samplers = mexdolphin$samplers,
domain = list(
geometry = mesh,
distance = fm_mesh_1d(seq(0, 8, length.out = 30))
),
formula = formula1
)Plots:
spde.range <- spde.posterior(fit1, "mySPDE", what = "range")
plot(spde.range)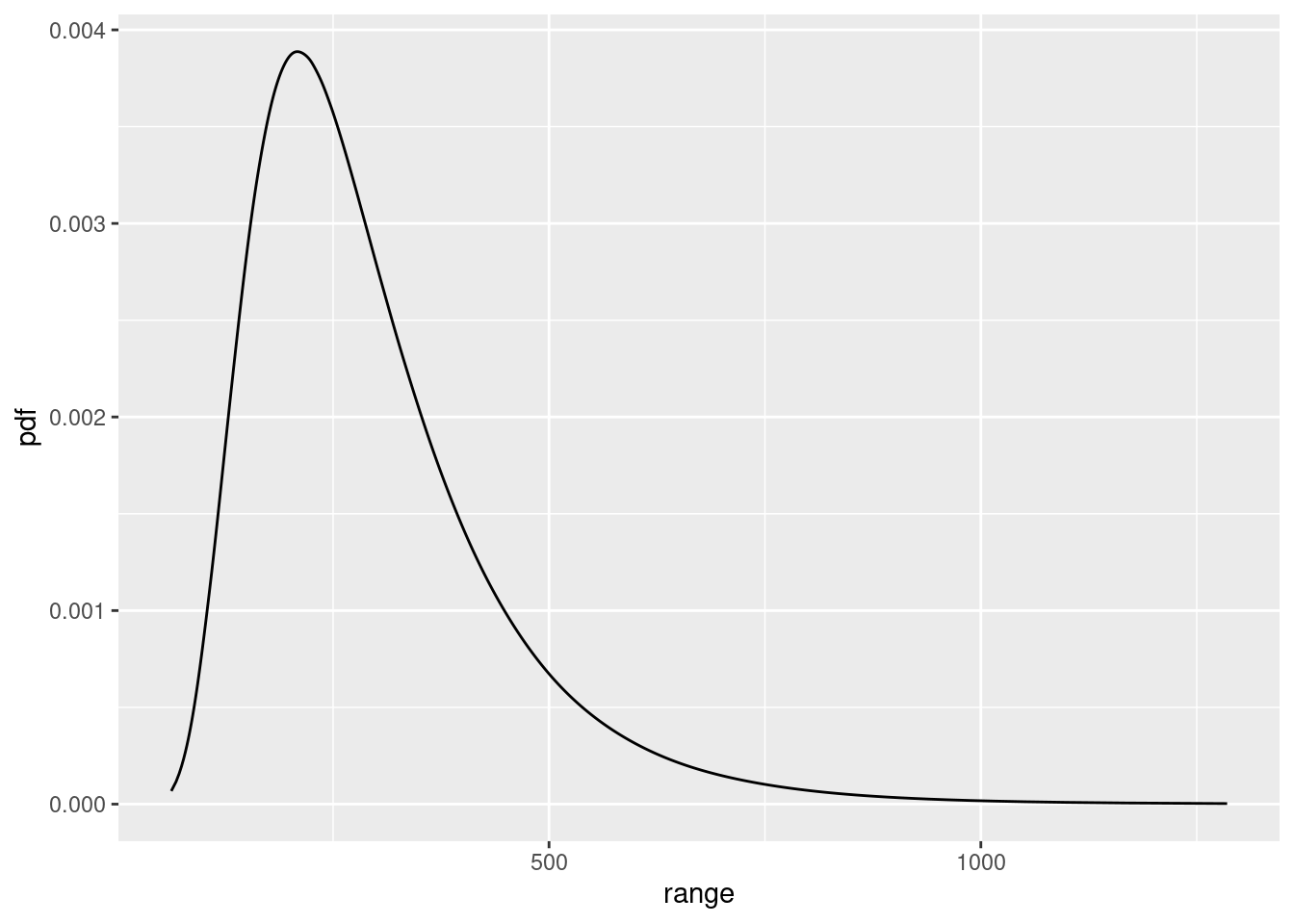
spde.logvar <- spde.posterior(fit1, "mySPDE", what = "log.variance")
plot(spde.logvar)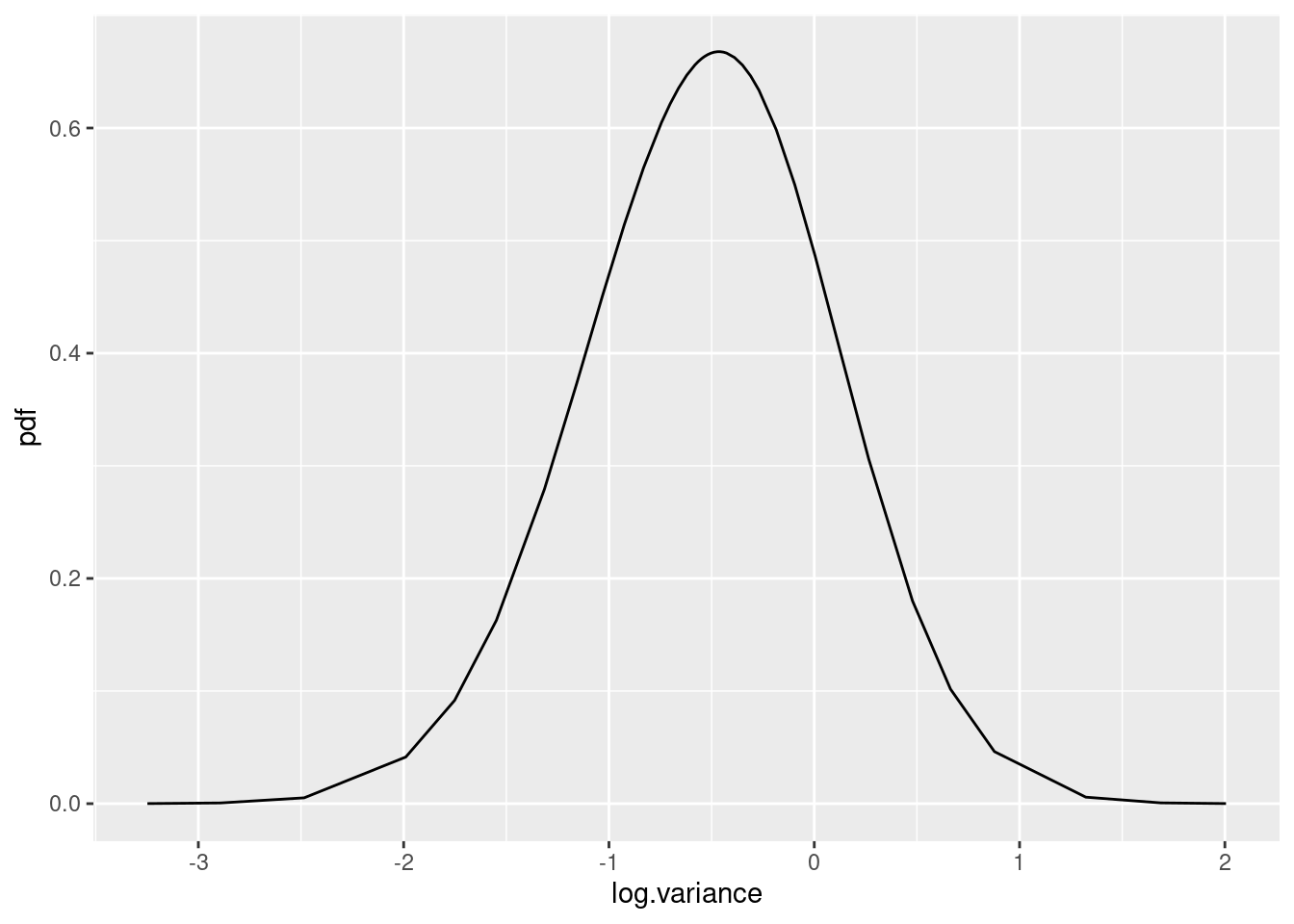
pr.int1 <- predict(fit1, pxl, ~ exp(mySPDE + Intercept))
ggplot() +
gg(pr.int1, geom = "tile") +
gg(mexdolphin$ppoly, linewidth = 1, alpha = 0) +
gg(mexdolphin$samplers, color = "grey") +
gg(mexdolphin$points, size = 0.2, alpha = 1) +
theme(
legend.key.width = unit(x = 0.2, "cm"),
legend.key.height = unit(x = 0.3, "cm")
) +
theme(legend.text = element_text(size = 6))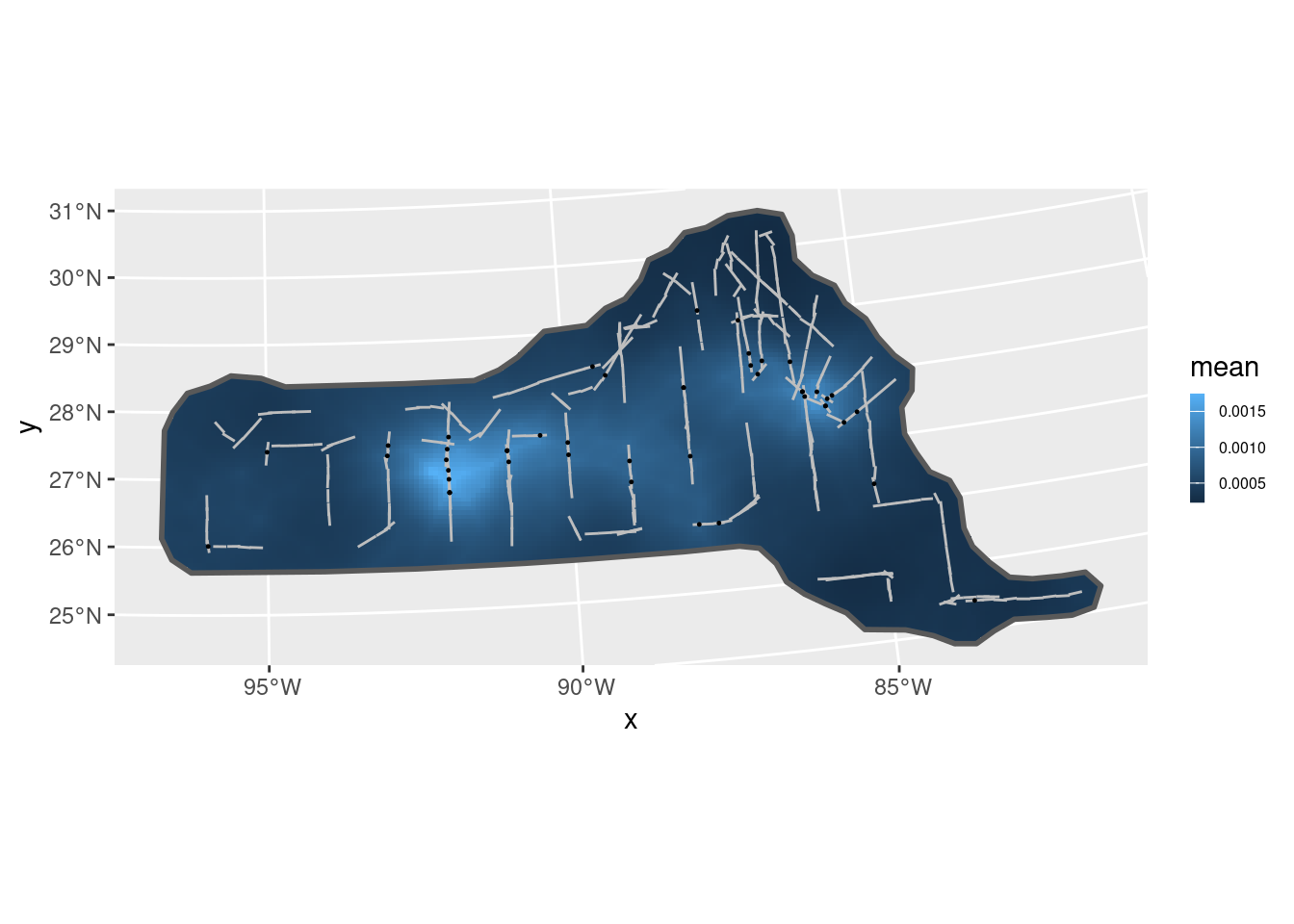
distdf <- data.frame(distance = seq(0, 8, length.out = 100))
dfun1 <- predict(fit1, distdf, ~ hr(distance, sigma))
plot(dfun1)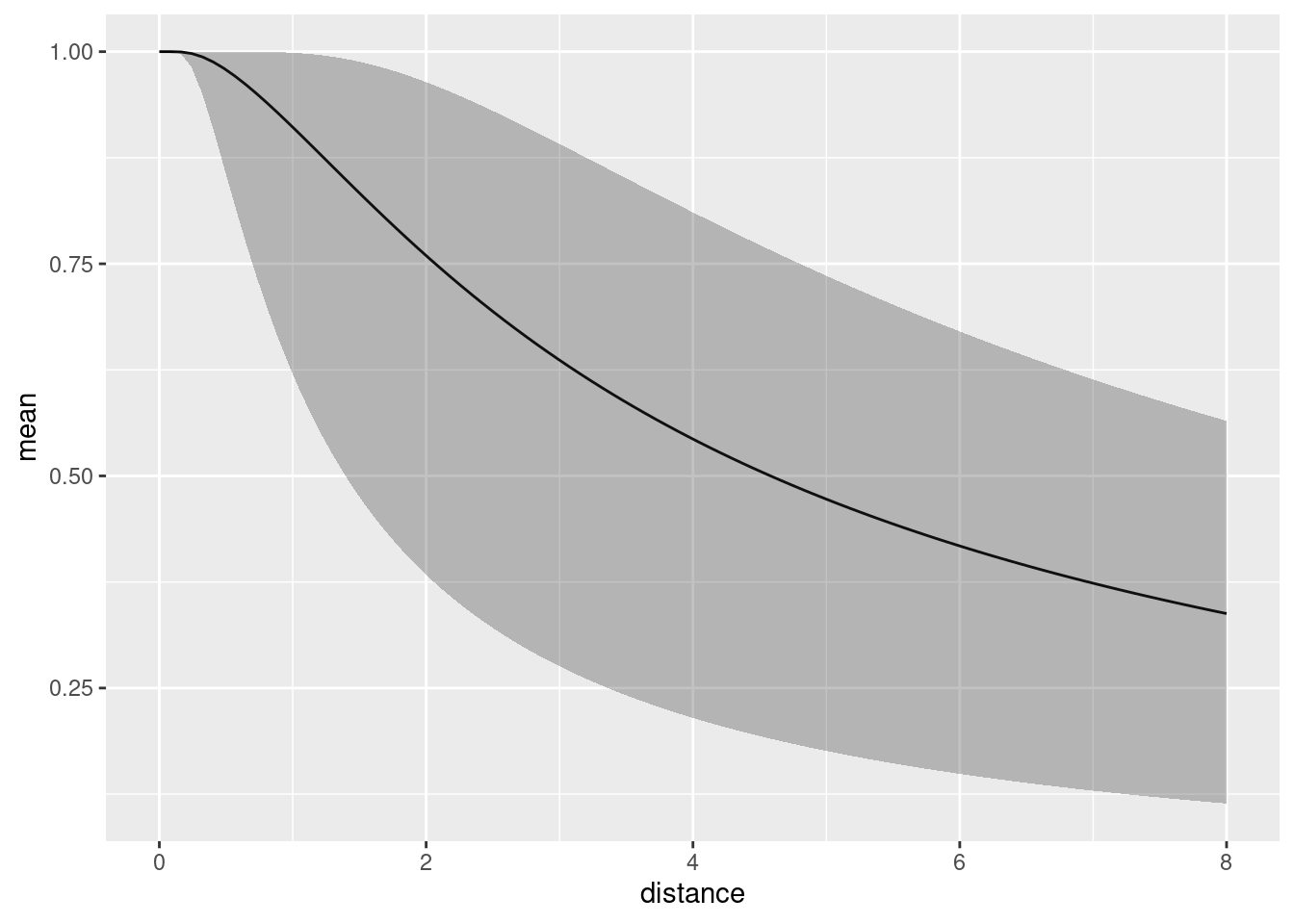
predpts <- fm_int(mexdolphin$mesh, mexdolphin$ppoly)
Lambda1 <- predict(fit1, predpts, ~ sum(weight * exp(mySPDE + Intercept)))
Lambda1
#> mean sd q0.025 q0.5 q0.975 median mean.mc_std_err
#> 1 295.6502 79.86857 168.9141 289.3827 467.5875 289.3827 9.074797
#> sd.mc_std_err
#> 1 5.439703
Ns <- seq(50, 650, by = 1)
Nest1 <- predict(
fit1,
predpts,
~ data.frame(
N = Ns,
density = dpois(
Ns,
lambda = sum(weight * exp(mySPDE + Intercept))
)
),
n.samples = 2000
)
Nest1 <- dplyr::bind_rows(
cbind(Nest1, Method = "Posterior"),
data.frame(
N = Nest1$N,
mean = dpois(Nest1$N, lambda = Lambda1$mean),
mean.mc_std_err = 0,
Method = "Plugin"
)
)
ggplot(data = Nest1) +
geom_line(aes(x = N, y = mean, colour = Method)) +
geom_ribbon(
aes(
x = N,
ymin = mean - 2 * mean.mc_std_err,
ymax = mean + 2 * mean.mc_std_err,
fill = Method
),
alpha = 0.2
) +
geom_line(aes(x = N, y = mean, colour = Method)) +
ylab("Probability mass function")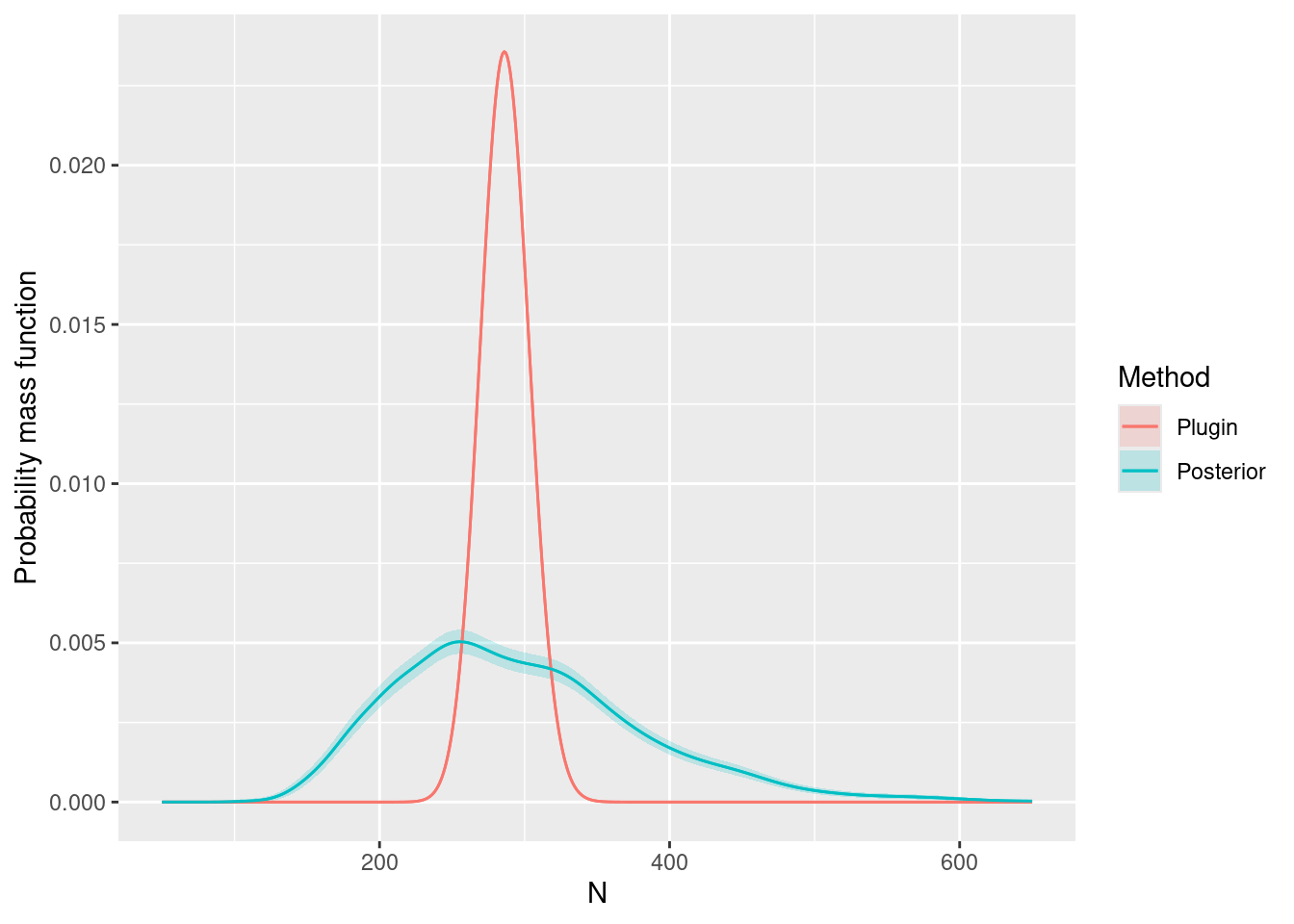
Comparing the models
Look at the goodness-of-fit of the two models in the distance dimension
bc <- bincount(
result = fit,
observations = mexdolphin$points$distance,
breaks = seq(0, max(mexdolphin$points$distance), length.out = 9),
predictor = distance ~ hn(distance, sigma)
)
attributes(bc)$ggp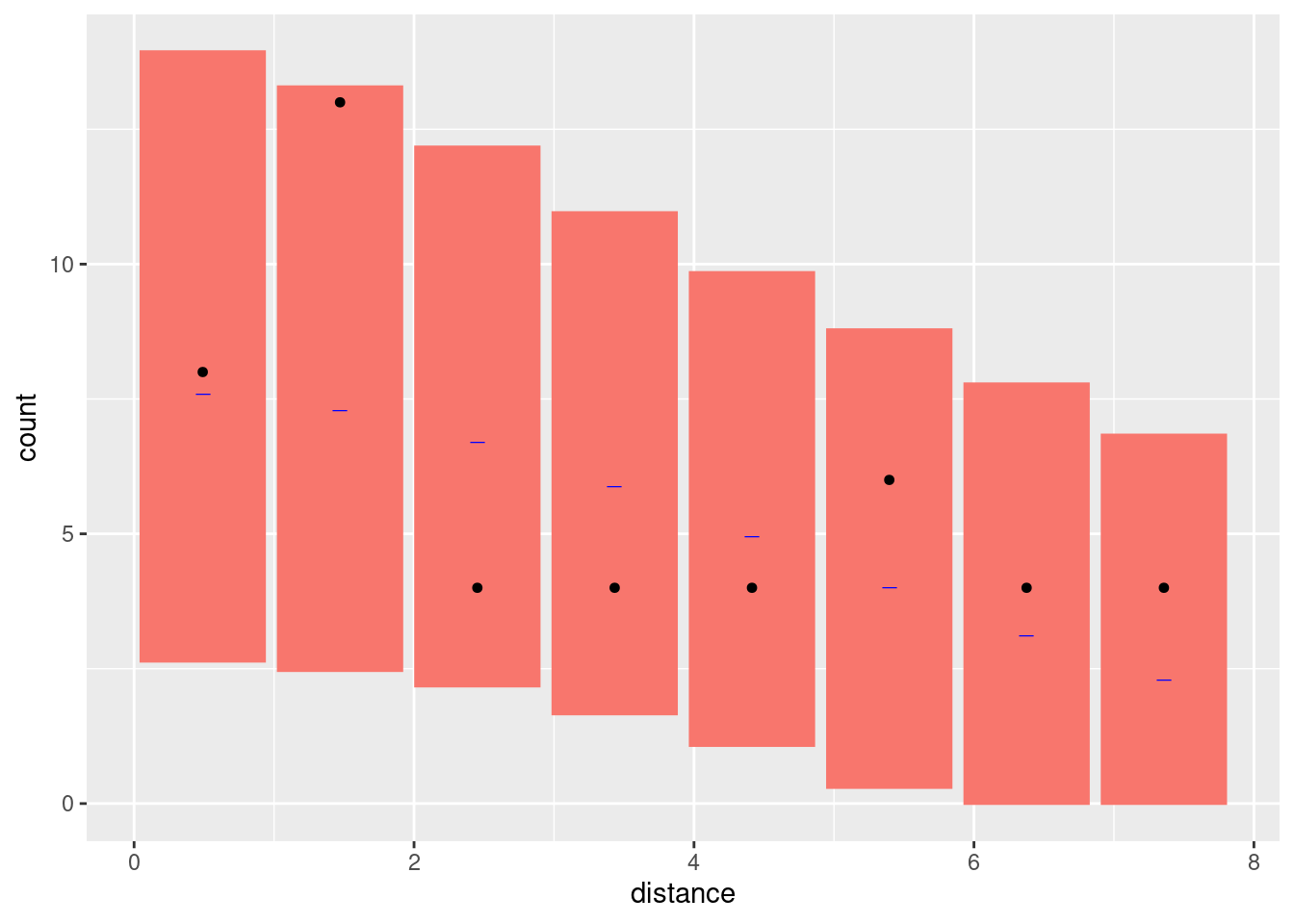
bc1 <- bincount(
result = fit1,
observations = mexdolphin$points$distance,
breaks = seq(0, max(mexdolphin$points$distance), length.out = 9),
predictor = distance ~ hn(distance, sigma)
)
attributes(bc1)$ggp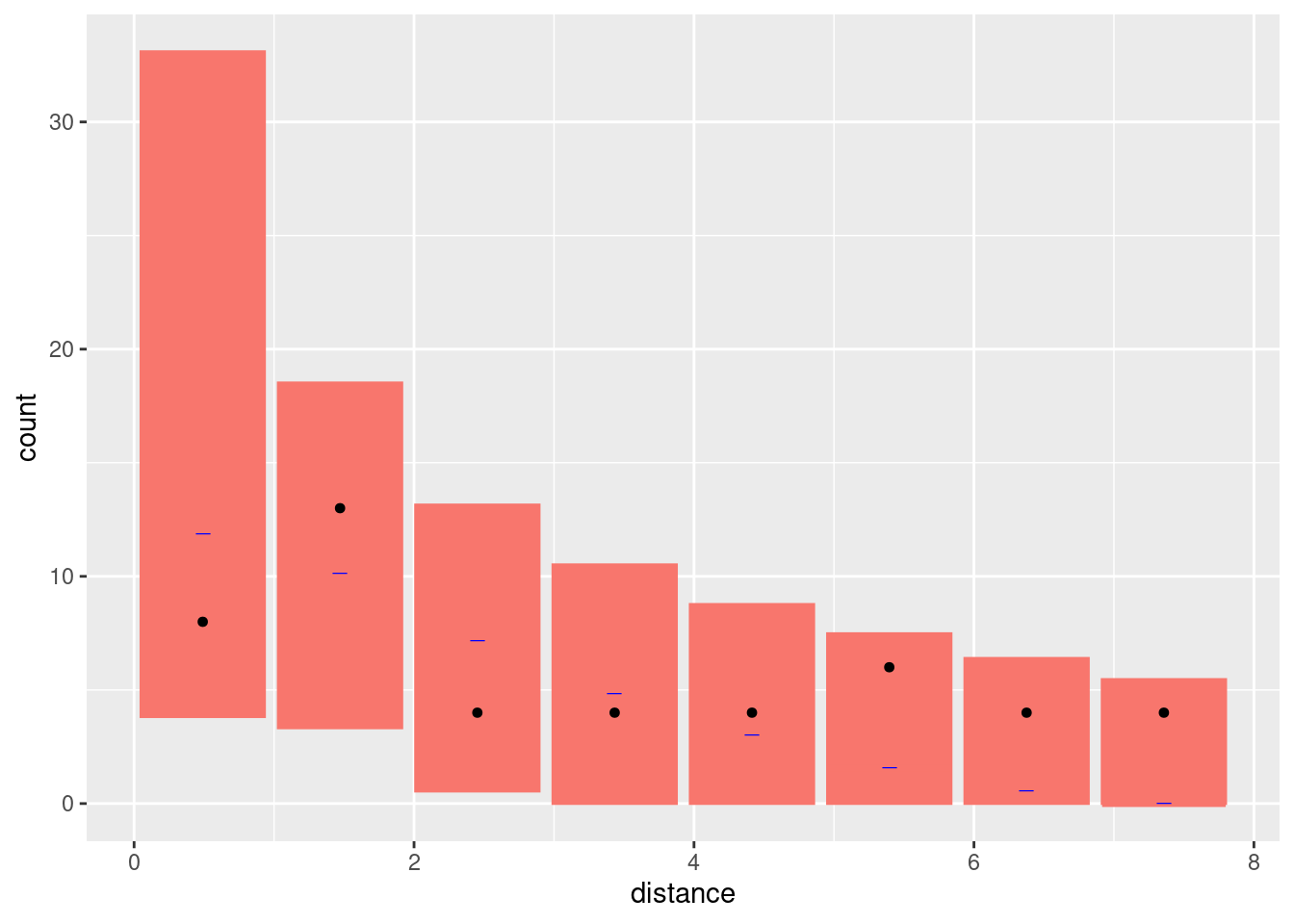
Fit Models only to the distance sampling data
Half-normal first
formula <- distance ~ log(hn(distance, sigma)) + Intercept
cmp <- ~ sigma(1,
prec.linear = 1,
marginal = bm_marginal(qexp, pexp, dexp, rate = 1 / 8)
) +
Intercept(1)
dfit <- lgcp(
components = cmp,
mexdolphin$points,
domain = list(distance = fm_mesh_1d(seq(0, 8, length.out = 30))),
formula = formula,
options = list(bru_initial = list(sigma = 1, Intercept = 3))
)
detfun <- predict(dfit, distdf, ~ hn(distance, sigma))Hazard-rate next
formula1 <- distance ~ log(hr(distance, sigma)) + Intercept
cmp <- ~ sigma(1,
prec.linear = 1,
marginal = bm_marginal(qexp, pexp, dexp, rate = 1 / 8)
) +
Intercept(1)
dfit1 <- lgcp(
components = cmp,
mexdolphin$points,
domain = list(distance = fm_mesh_1d(seq(0, 8, length.out = 30))),
formula = formula1
)
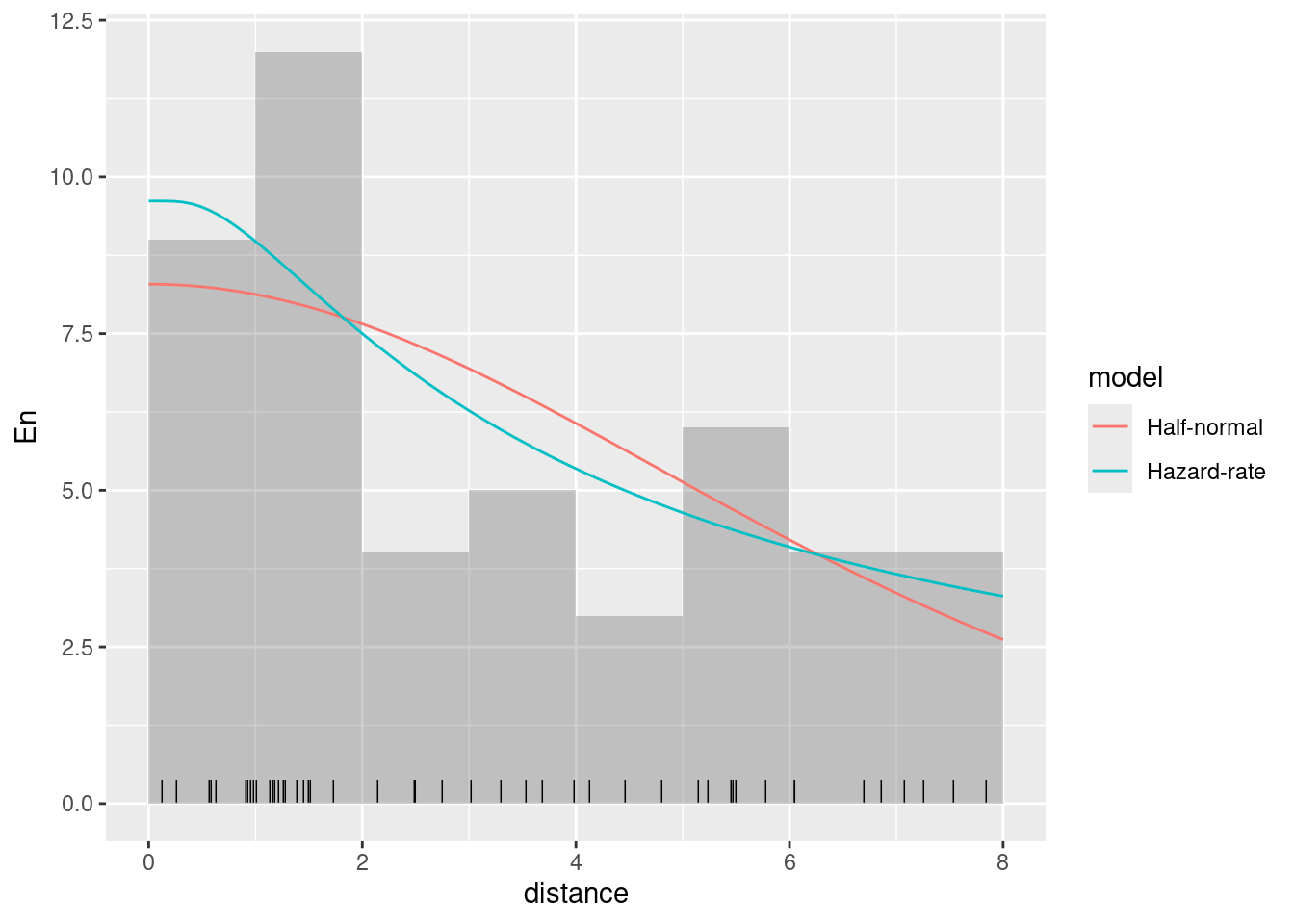
detfun1 <- predict(dfit1, distdf, ~ hr(distance, sigma))Plot both lines on histogram of observations. First scale lines to have same area as that of histogram.
Half-normal:
hnline <- data.frame(
distance = detfun$distance,
p = detfun$mean,
lower = detfun$q0.025,
upper = detfun$q0.975
)
wts <- diff(hnline$distance)
wts[1] <- wts[1] / 2
wts <- c(wts, wts[1])
hnarea <- sum(wts * hnline$p)
n <- length(mexdolphin$points$distance)
scale <- n / hnarea
hnline$En <- hnline$p * scale
hnline$En.lower <- hnline$lower * scale
hnline$En.upper <- hnline$upper * scaleHazard-rate:
hrline <- data.frame(
distance = detfun1$distance,
p = detfun1$mean,
lower = detfun1$q0.025,
upper = detfun1$q0.975
)
wts <- diff(hrline$distance)
wts[1] <- wts[1] / 2
wts <- c(wts, wts[1])
hrarea <- sum(wts * hrline$p)
n <- length(mexdolphin$points$distance)
scale <- n / hrarea
hrline$En <- hrline$p * scale
hrline$En.lower <- hrline$lower * scale
hrline$En.upper <- hrline$upper * scaleCombine lines in a single object for plotting
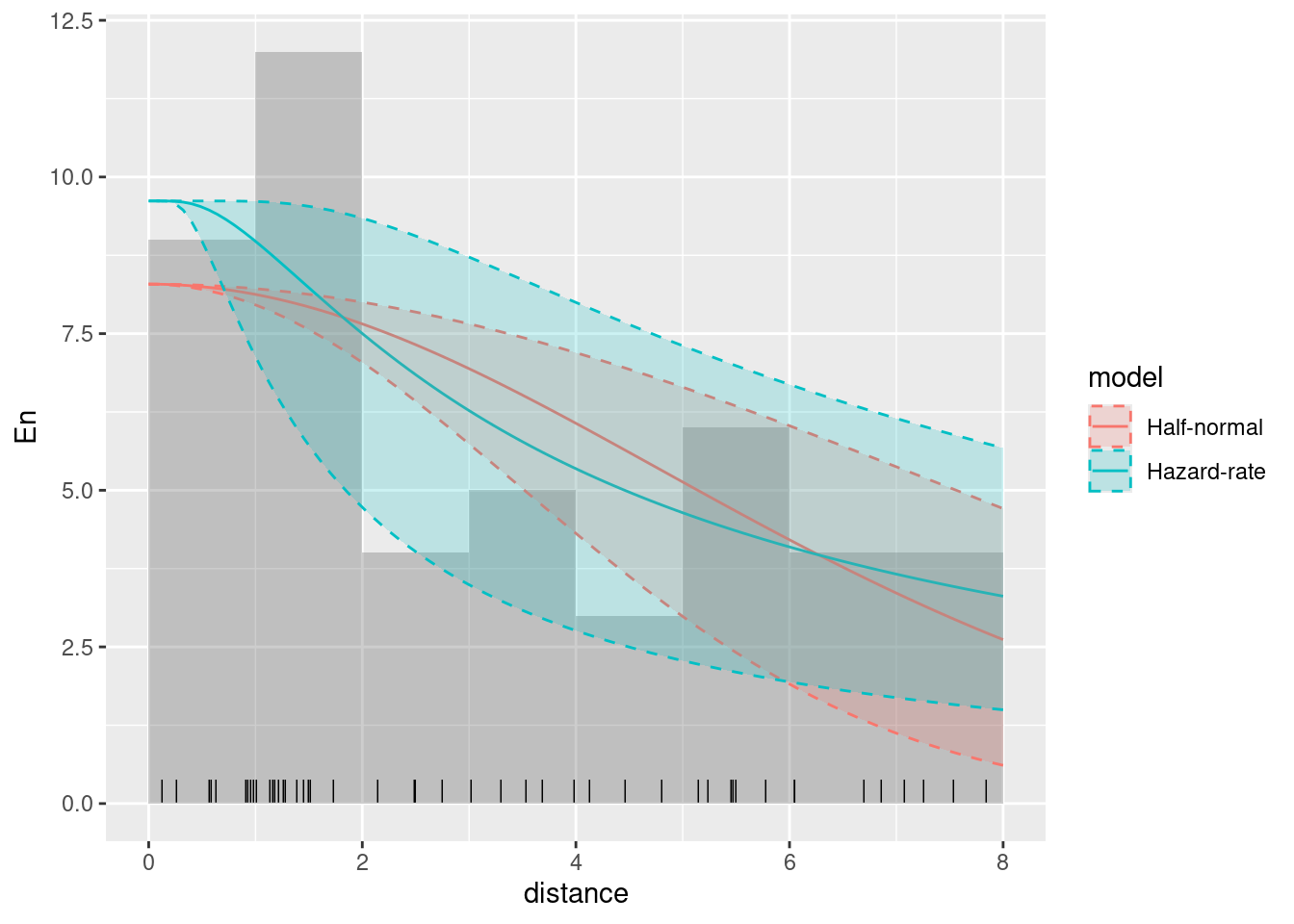
Plot without the 95% credible intervals
ggplot(data.frame(mexdolphin$points)) +
geom_histogram(aes(x = distance),
breaks = seq(0, 8, length.out = 9),
alpha = 0.3
) +
geom_point(aes(x = distance), y = 0.2, shape = "|", size = 3) +
geom_line(
data = dlines,
aes(x = distance, y = En, group = model, col = model)
)
Plot with the 95% credible intervals (without taking the count rescaling into account)
ggplot(data.frame(mexdolphin$points)) +
geom_histogram(aes(x = distance),
breaks = seq(0, 8, length.out = 9),
alpha = 0.3
) +
geom_point(aes(x = distance), y = 0.2, shape = "|", size = 3) +
geom_line(
data = dlines,
aes(x = distance, y = En, group = model, col = model)
) +
geom_ribbon(
data = dlines,
aes(
x = distance,
ymin = En.lower,
ymax = En.upper,
group = model,
col = model,
fill = model
),
alpha = 0.2, lty = 2
)