LGCPs - Plot sampling
David Borchers and Finn Lindgren
Generated on 2026-02-26
Source:vignettes/articles/2d_lgcp_plotsampling.Rmd
2d_lgcp_plotsampling.RmdIntroduction
This practical demonstrates use of the samplers argument
in lgcp, which you need to use when you have observed
points from only a sample of plots in the survey region.
Get the data
data(gorillas_sf, package = "inlabru")This dataset is a list (see help(gorillas_sf) for
details. Extract the the objects you need from the list, for
convenience:
nests <- gorillas_sf$nests
mesh <- gorillas_sf$mesh
boundary <- gorillas_sf$boundary
gcov <- gorillas_sf_gcov()
#> Loading required namespace: terraThe gorillas_sf data also contains a plot sample subset
which covers 60% of the survey region.
sample <- gorillas_sf$plotsample
plotdets <- ggplot() +
gg(boundary) +
gg(sample$plots) +
gg(sample$nests, pch = "+", cex = 4, color = "red") +
geom_text(aes(
label = sample$counts$count,
x = sf::st_coordinates(sample$counts)[, 1],
y = sf::st_coordinates(sample$counts)[, 2]
)) +
labs(x = "Easting", y = "Northing")
plot(plotdets)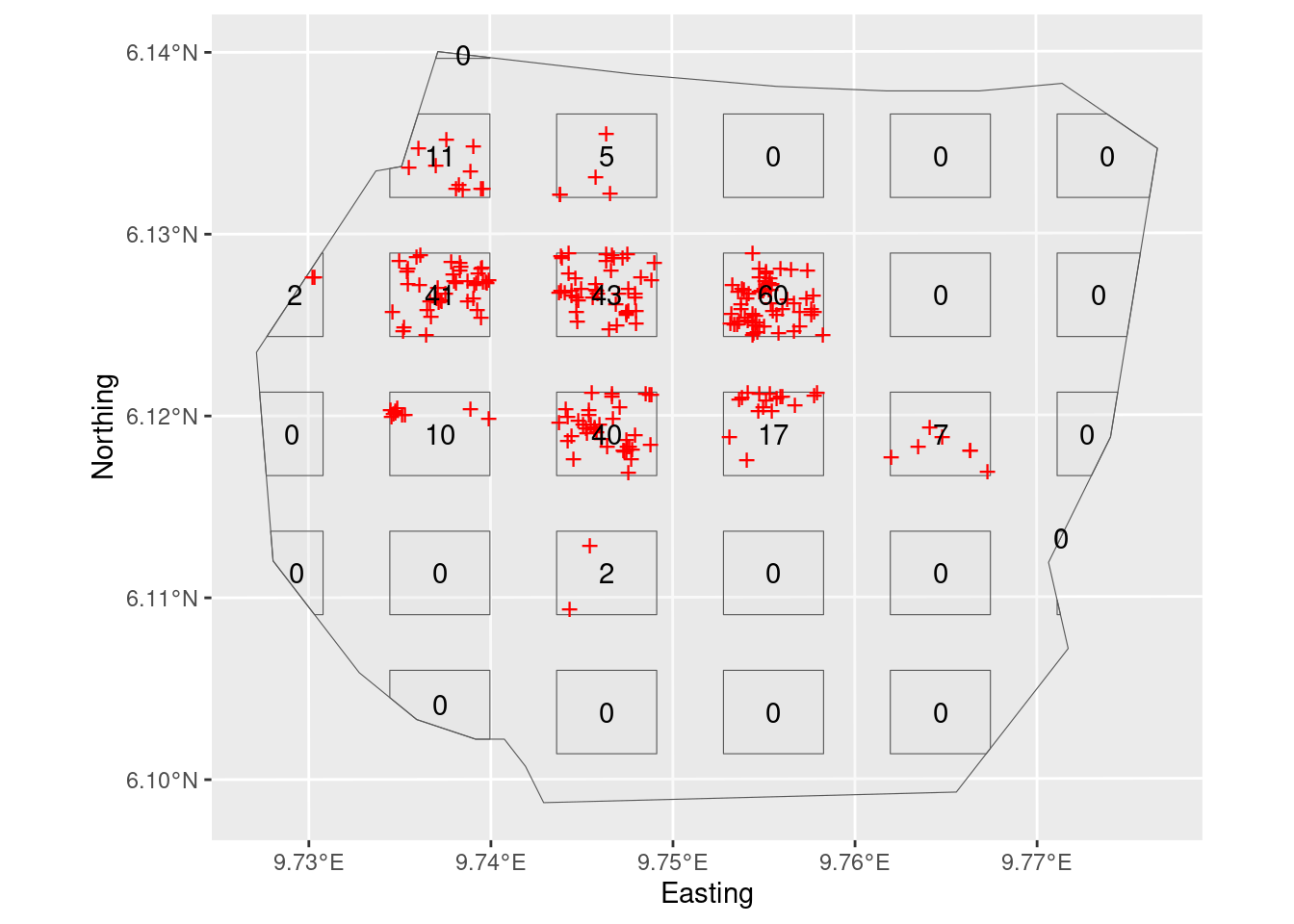
On this plot survey, only points within the rectangles are detected, but it is also informative to plot all the points here (which if it was a real plot survey you could not do, because you would not have seen them all).
plotwithall <- ggplot() +
gg(boundary) +
gg(sample$plots) +
gg(nests, pch = "+", cex = 4, color = "blue") +
geom_text(aes(
label = sample$counts$count,
x = sf::st_coordinates(sample$counts)[, 1],
y = sf::st_coordinates(sample$counts)[, 2]
)) +
gg(sample$nests, pch = "+", cex = 4, color = "red") +
labs(x = "Easting", y = "Northing")
plot(plotwithall)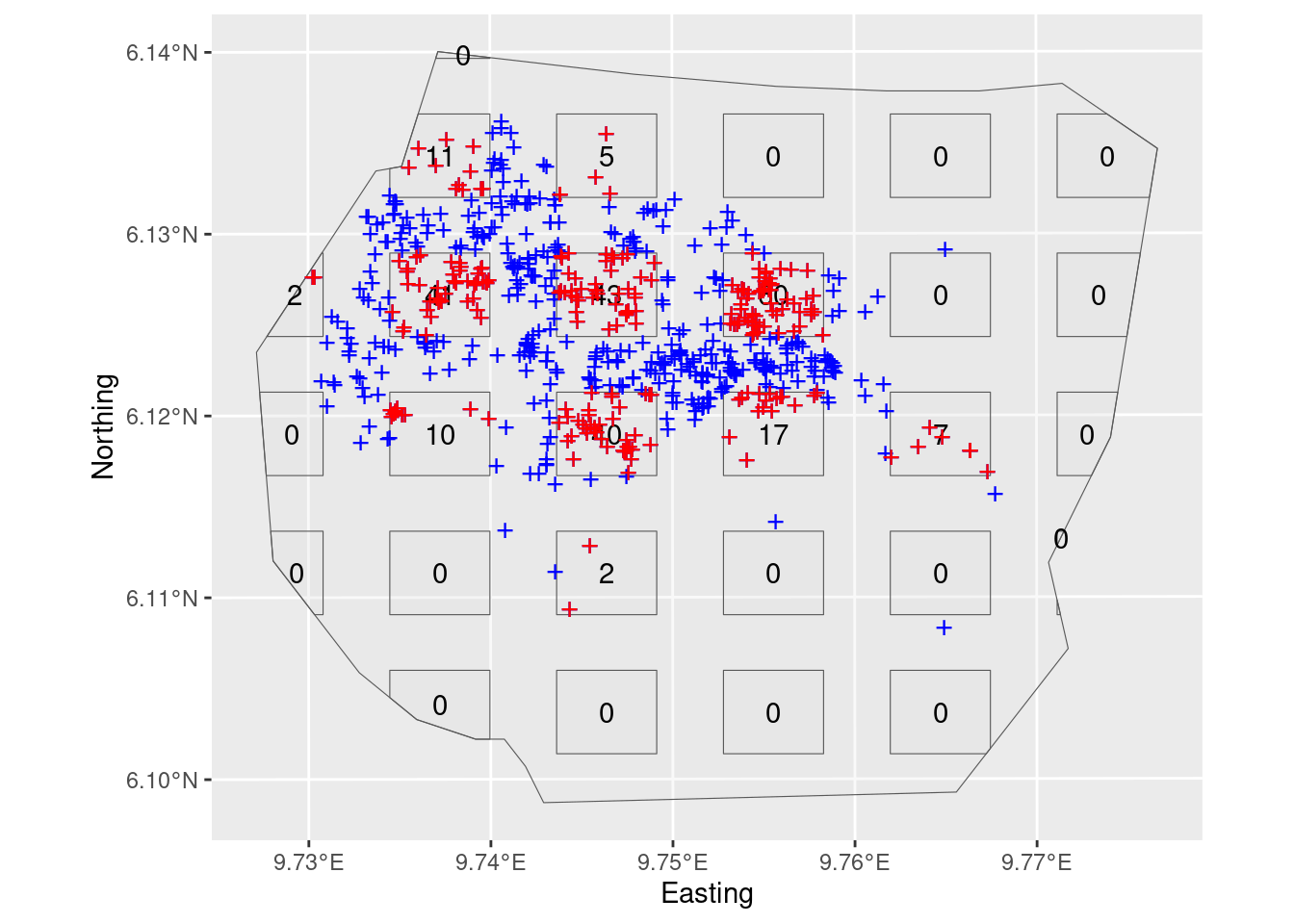
Inference
The observed nest locations are in the sf
sample$nests, and the plots are in the sf
sample$plots. Again, we are using the following SPDE
setup:
matern <- inla.spde2.pcmatern(mesh,
prior.sigma = c(0.1, 0.01),
prior.range = c(0.05, 0.01)
)Fit an LGCP model with SPDE only to these data by using the
samplers= argument of the function
lgcp( ):
cmp <- geometry ~ my.spde(geometry, model = matern)
fit <- lgcp(cmp,
sample$nests,
samplers = sample$plots,
domain = list(geometry = mesh)
)Plot the density surface from your fitted model
pxl <- fm_pixels(mesh, mask = boundary)
lambda.sample <- predict(fit, pxl, ~ exp(my.spde + Intercept))
lambda.sample.plot <- ggplot() +
gg(lambda.sample, geom = "tile") +
gg(sample$plots, alpha = 0) +
gg(boundary, col = "yellow", alpha = 0)
lambda.sample.plot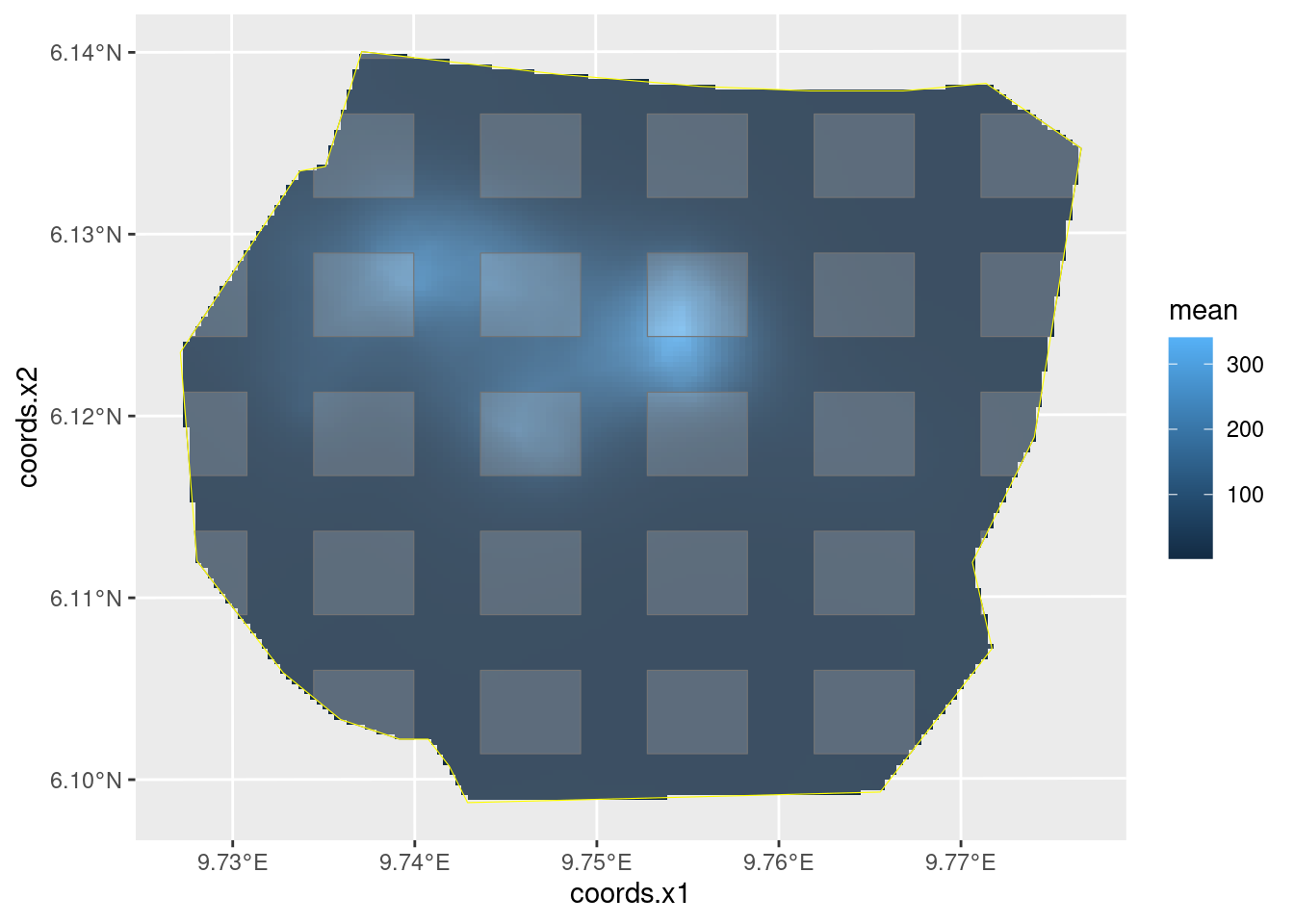
Estimate the integrated intensity lambda. We compute both the overall integrated intensity, representative of an imagined new realisation of the point process, and the conditional expectation that takes the actually observed nests into account, by recognising that we have complete information in the surveyed plots.
Lambda <- predict(
fit,
fm_int(mesh, boundary),
~ sum(weight * exp(my.spde + Intercept))
)
Lambda.empirical <- predict(
fit,
rbind(
cbind(fm_int(mesh, boundary), data.frame(all = TRUE)),
cbind(fm_int(mesh, sample$plots), data.frame(all = FALSE))
),
~ (sum(weight * exp(my.spde + Intercept) * all) -
sum(weight * exp(my.spde + Intercept) * !all) +
nrow(sample$nests))
)
rbind(
Lambda,
Lambda.empirical
)Fit the same model to the full dataset (the points in
gorillas_sf$nests), or get your previous fit, if you kept
it. Plot the intensity surface and estimate the integrated intensity
fit.all <- lgcp(cmp, nests,
samplers = boundary,
domain = list(geometry = mesh)
)
lambda.all <- predict(fit.all, pxl, ~ exp(my.spde + Intercept))
Lambda.all <- predict(
fit.all,
fm_int(mesh, boundary),
~ sum(weight * exp(my.spde + Intercept))
)Your plot should look like this:
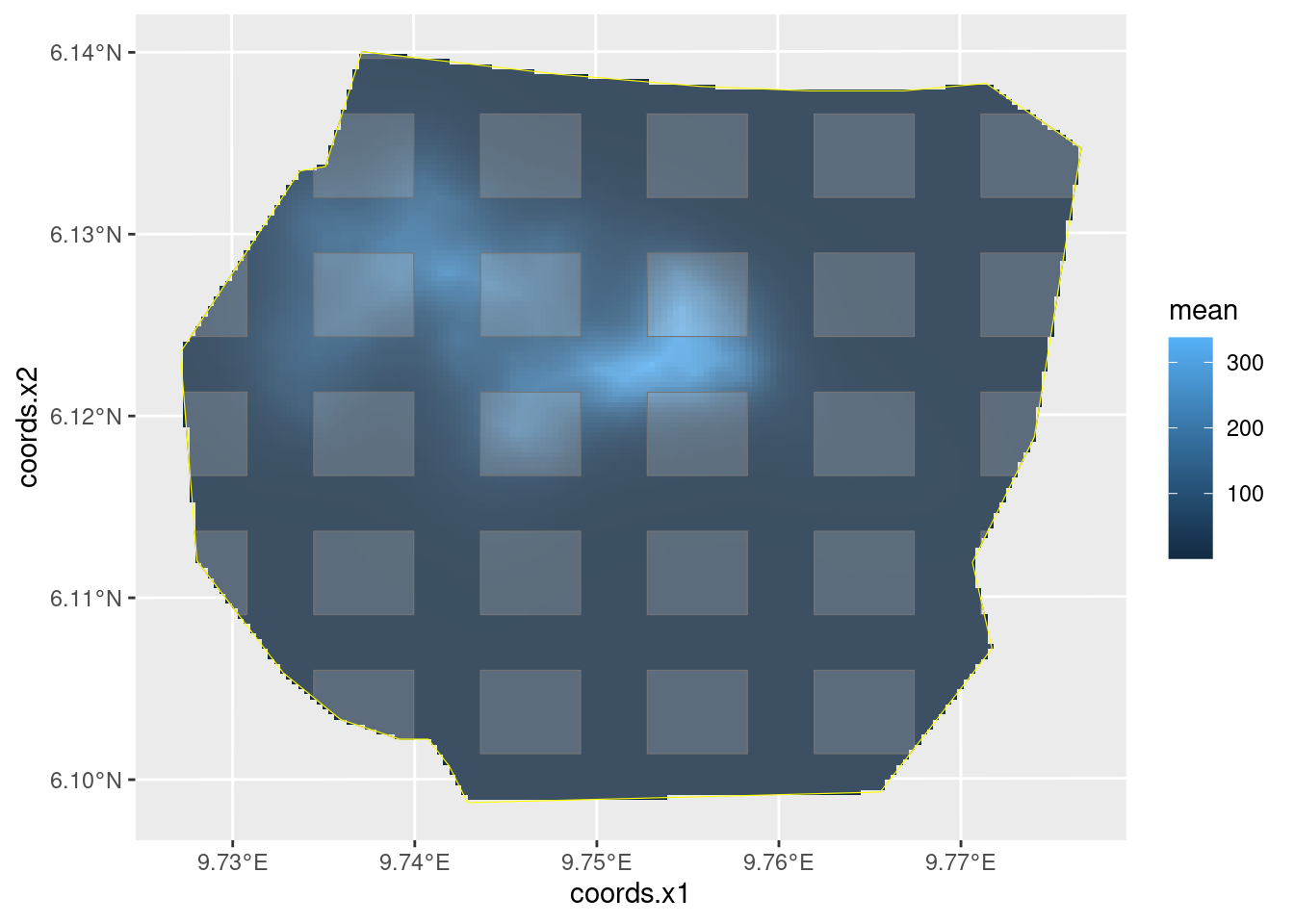
The values Lambda.empirical, Lambda, and
Lambda.all should be close to each other if the plot
samples gave sufficient information for the overall prediction:
rbind(
Plots = Lambda,
PlotsEmp = Lambda.empirical,
All = Lambda.all,
AllEmp = c(
nrow(gorillas_sf$nests),
0,
rep(nrow(gorillas_sf$nests), 3),
rep(NA, 3)
)
)
#> mean sd q0.025 q0.5 q0.975 median mean.mc_std_err
#> Plots 650.2641 45.59430 553.6804 653.2822 733.0771 653.2822 5.222740
#> PlotsEmp 646.4900 42.34880 576.6405 639.8072 737.3688 639.8072 4.817688
#> All 672.5919 25.42693 628.5485 673.7731 727.0829 673.7731 2.888124
#> AllEmp 647.0000 0.00000 647.0000 647.0000 647.0000 NA NA
#> sd.mc_std_err
#> Plots 3.316552
#> PlotsEmp 2.914038
#> All 1.727155
#> AllEmp NANow, let’s compare the results
library(patchwork)
lambda.sample.plot + lambda.all.plot +
plot_layout(guides = "collect") &
theme(legend.position = "left") &
scale_fill_continuous(limits = range(c(0, 340)))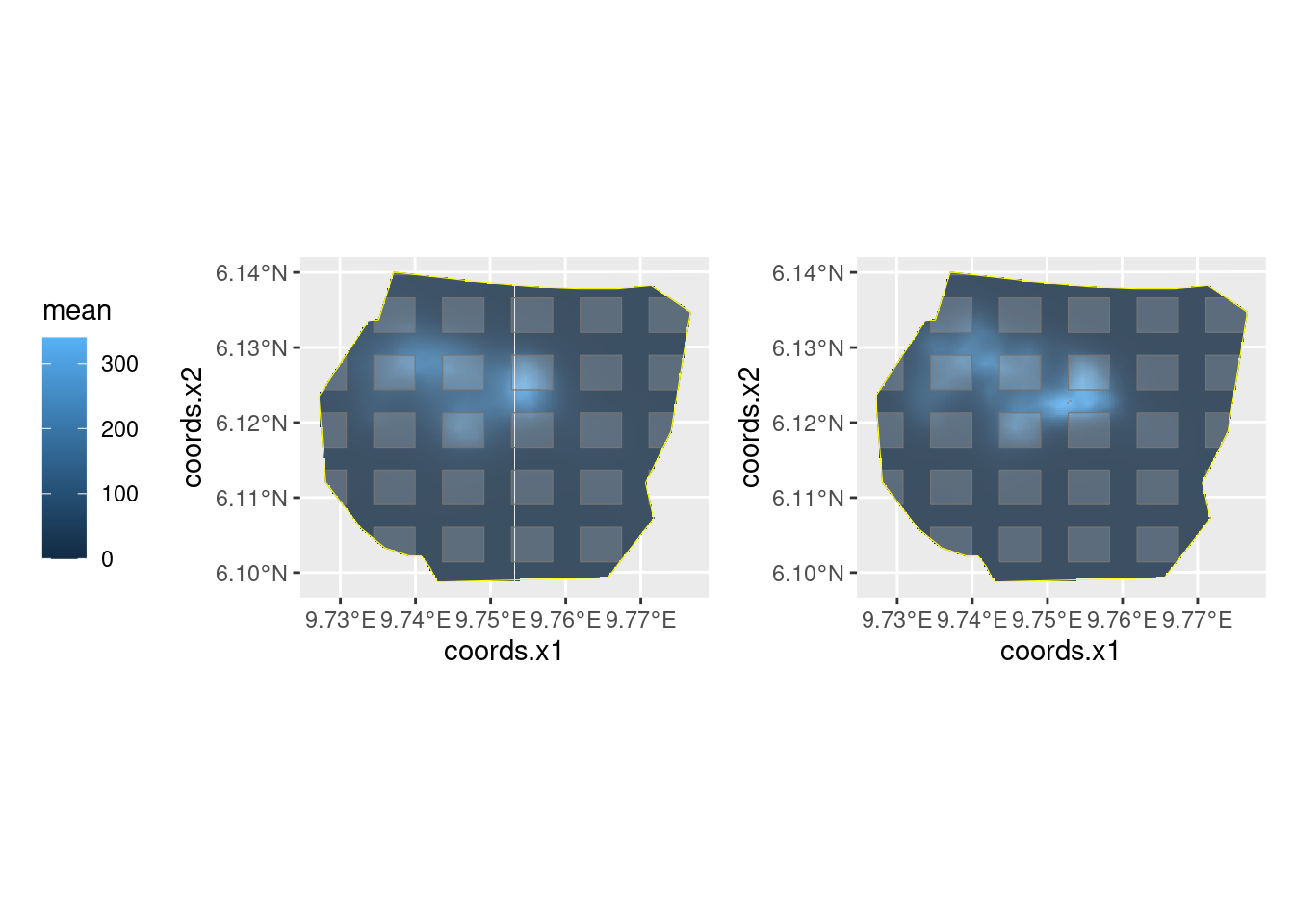
Do you understand the reason for the differences in the posteriors of the abundance estimates?