Generic function for sampling for fitted models. The function invokes particular methods which depend on the class of the first argument.
Takes a fitted bru object produced by the function bru() and produces
samples given a new set of values for the model covariates or the original
values used for the model fit. The samples can be based on any R expression
that is valid given these values/covariates and the joint
posterior of the estimated random effects.
Usage
generate(object, ...)
# S3 method for class 'bru'
generate(
object,
newdata = NULL,
formula = NULL,
n.samples = 100,
seed = 0L,
num.threads = NULL,
used = NULL,
...,
data = deprecated(),
include = deprecated(),
exclude = deprecated()
)Arguments
- object
A
bruobject obtained by callingbru().- ...
additional, unused arguments.
- newdata
A
data.frameorSpatialPointsDataFrameof covariates needed for sampling.- formula
A formula where the right hand side defines an R expression to evaluate for each generated sample. If
NULL, the latent and hyperparameter states are returned as named list elements. See Details for more information.- n.samples
Integer setting the number of samples to draw in order to calculate the posterior statistics. The default, 100, is rather low but provides a quick approximate result.
- seed
Random number generator seed passed on to
INLA::inla.posterior.sample- num.threads
Specification of desired number of threads for parallel computations. Default NULL, leaves it up to INLA. When seed != 0, overridden to "1:1"
- used
Either
NULLor abru_used()object. Default,NULL, uses auto-detection of used variables in the formula.- data
- include, exclude
If auto-detection of used variables fails, use
usedinstead.
Value
The form of the value returned by generate() depends on the data
class and prediction formula. Normally, a data.frame is returned, or a list
of data.frames (if the prediction formula generates a list)
List of generated samples
Details
In addition to the component names (that give the effect of each component
evaluated for the input data), the suffix _latent variable name can be used
to directly access the latent state for a component, and the suffix function
_eval can be used to evaluate a component at other input values than the
expressions defined in the component definition itself, e.g.
field_eval(cbind(x, y)) for a component that was defined with
field(coordinates, ...) (see also bru_comp_eval()).
For "iid" models with mapper = bm_index(n), rnorm() is used to
generate new realisations for indices greater than n, if accessed
via <name>_eval(...).
Examples
# \donttest{
if (bru_safe_inla() &&
require("sn", quietly = TRUE)) {
# Generate data for a simple linear model
input.df <- data.frame(x = cos(1:10))
input.df <- within(
input.df,
y <- 5 + 2 * cos(1:10) + rnorm(10, mean = 0, sd = 0.1)
)
# Fit the model
fit <- bru(y ~ xeff(main = x, model = "linear"),
family = "gaussian", data = input.df
)
summary(fit)
# Generate samples for some predefined x
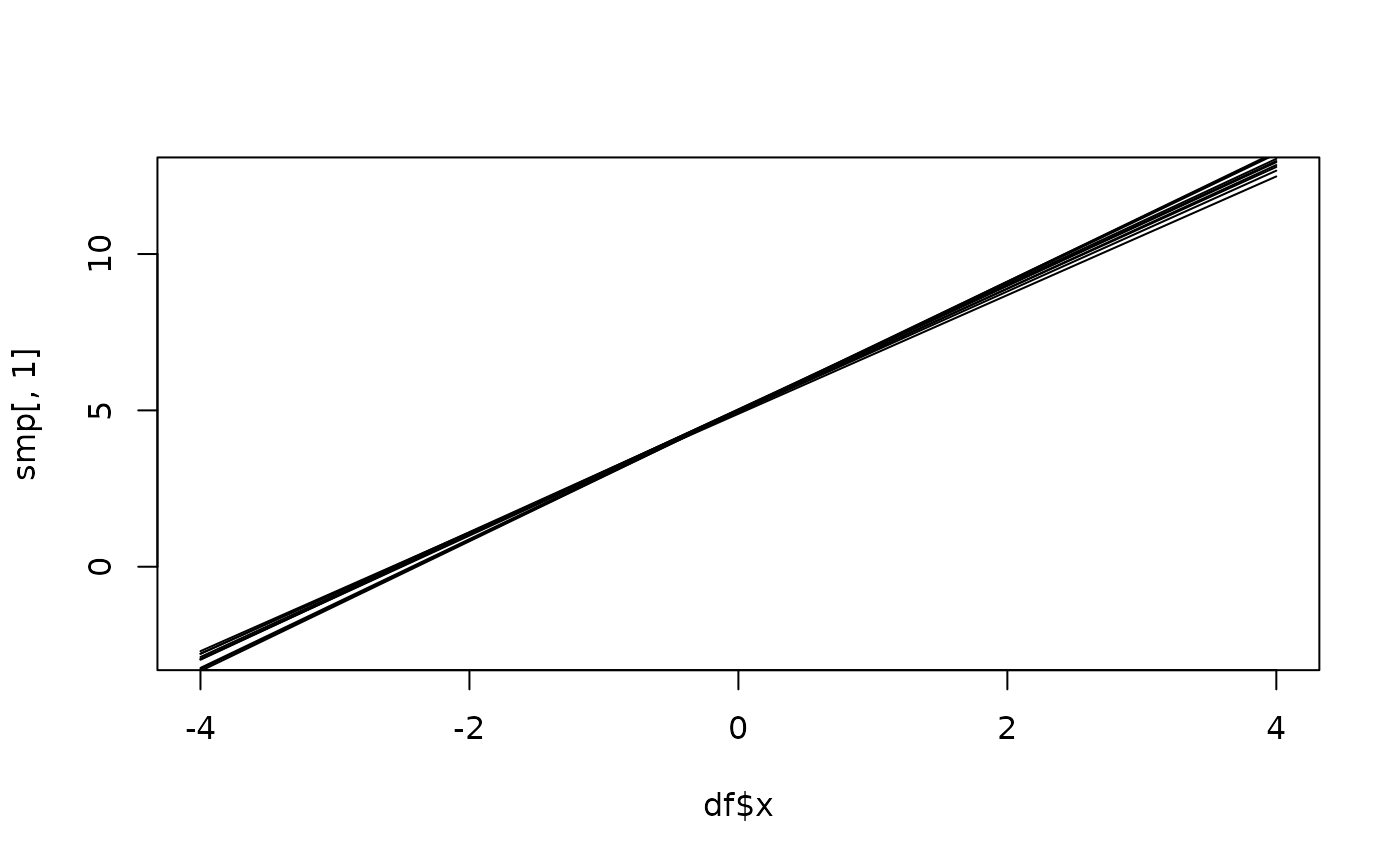
df <- data.frame(x = seq(-4, 4, by = 0.1))
smp <- generate(fit, df, ~ xeff + Intercept, n.samples = 10)
# Plot the resulting realizations
plot(df$x, smp[, 1], type = "l")
for (k in 2:ncol(smp)) points(df$x, smp[, k], type = "l")
# We can also draw samples form the joint posterior
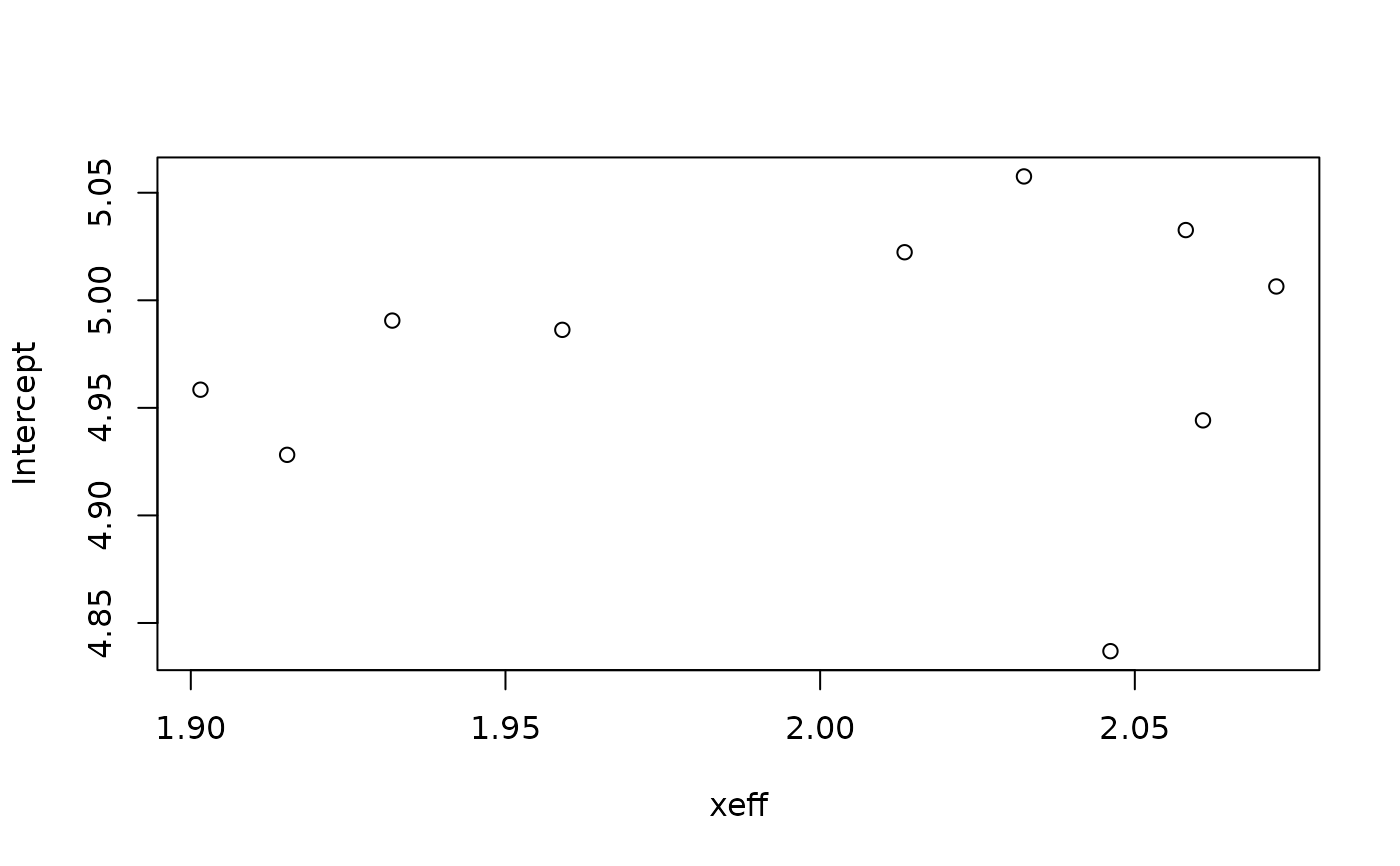
df <- data.frame(x = 1)
smp <- generate(fit, df, ~ data.frame(xeff, Intercept), n.samples = 10)
smp[[1]]
# ... and plot them
if (require(ggplot2, quietly = TRUE)) {
plot(do.call(rbind, smp))
}
}
 # }
# }