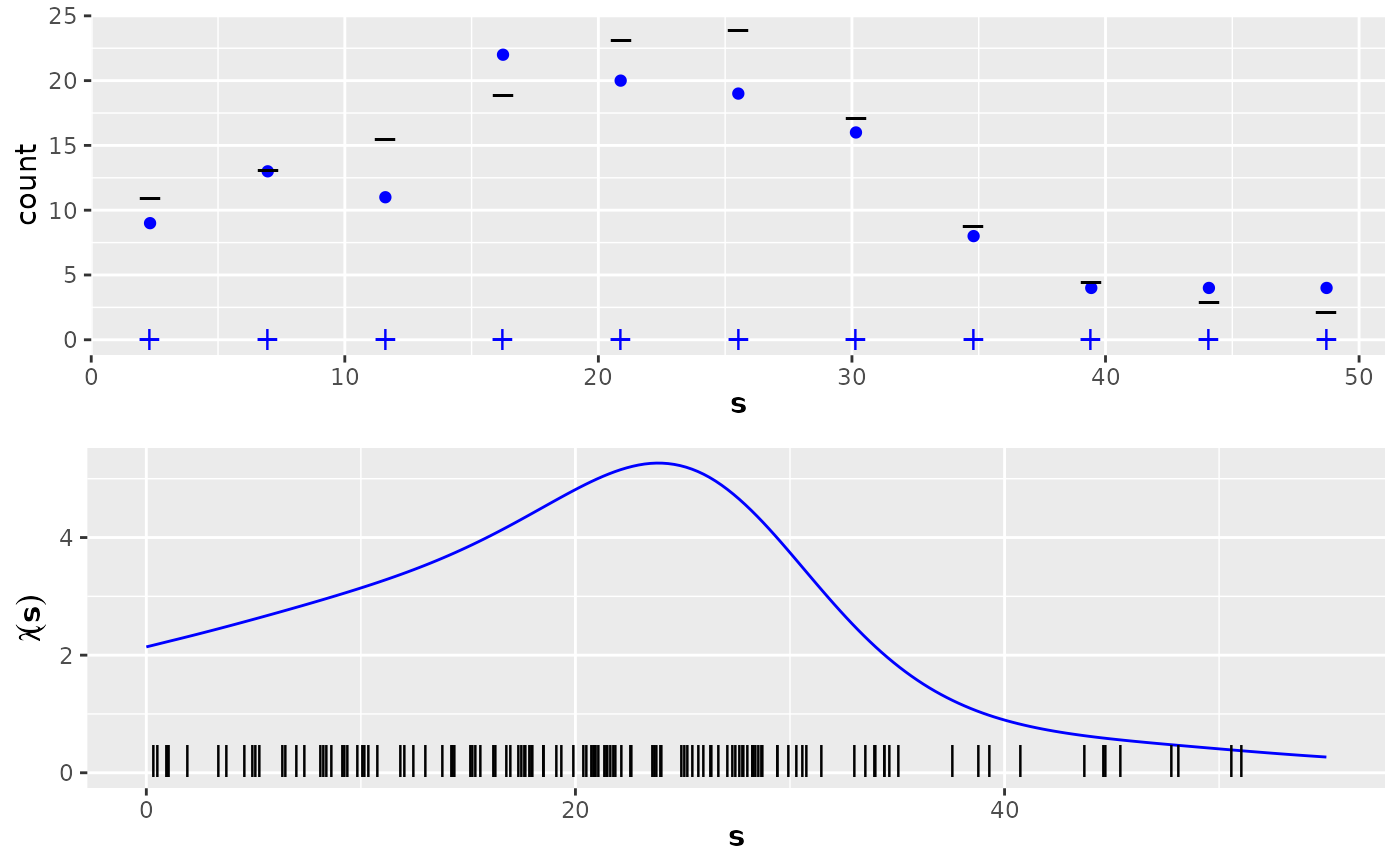
Point data and count data, together with intensity function and expected counts for a unimodal nonhomogeneous 1-dimensional Poisson process example.
Usage
data(Poisson2_1D)Format
The data contain the following R objects:
lambda2_1D:A function defining the intensity function of a nonhomogeneous Poisson process. Note that this function is only defined on the interval (0,55).
cov2_1D:A function that gives what we will call a 'habitat suitability' covariate in 1D space.
E_nc2The expected counts of the gridded data.
pts2The locations of the observed points (a data frame with one column, named
x).countdata2A data frame with three columns, containing the count data:
Examples
# \donttest{
if (require("ggplot2", quietly = TRUE) &&
require("patchwork", quietly = TRUE)) {
data(Poisson2_1D)
p1 <- ggplot(countdata2) +
geom_point(data = countdata2, aes(x = x, y = count), col = "blue") +
ylim(0, max(countdata2$count, E_nc2)) +
geom_point(
data = countdata2, aes(x = x), y = 0, shape = "+",
col = "blue", cex = 4
) +
geom_point(
data = data.frame(x = countdata2$x, y = E_nc2), aes(x = x),
y = E_nc2, shape = "_", cex = 5
) +
xlab(expression(bold(s))) +
ylab("count")
ss <- seq(0, 55, length.out = 200)
lambda <- lambda2_1D(ss)
p2 <- ggplot() +
geom_line(
data = data.frame(x = ss, y = lambda),
aes(x = x, y = y), col = "blue"
) +
ylim(0, max(lambda)) +
geom_point(data = pts2, aes(x = x), y = 0.2, shape = "|", cex = 4) +
xlab(expression(bold(s))) +
ylab(expression(lambda(bold(s))))
(p1 / p2)
}
 # }
# }