The Integrated nested Laplace approximation for fitting Dirichlet regression models
Joaquin Martinez-Minaya
2024-11-21
Source:vignettes/articles/example.Rmd
example.RmdIntroduction
This vignette is devoted to explain how to use the package dirinla. It is a R-package to fit Dirichlet regression models using R-INLA. It can be installed and upgraded via the repository https://github.com/inlabru-org/dirinla. In this manual, we simulate some data from a Dirichlet distribution to posteriorly fit them using the main function of the package dirinla.
Simulation
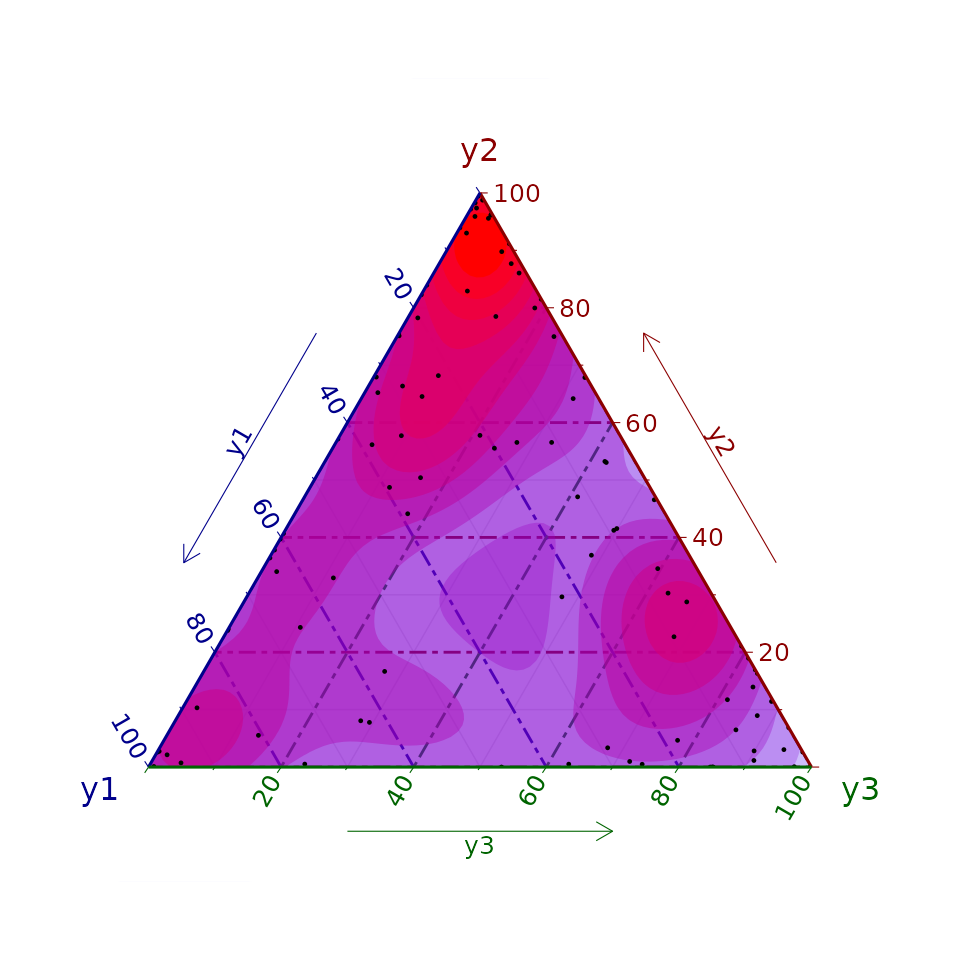
We firstly illustrate how to simulate 100 data points from a Dirichlet regression model with three different categories and one different covariate per category: being the parameters that compose the latent field , , (the intercepts), and , , (the slopes). Note that covariates are different for each category. This could be particularized for a situation where all of them are the same. For simplicity, covariates are simulated from a Uniform distribution on (0,1). To posteriorly fit the model, a following the structure of LGMs, Gaussian prior distributions are assigned with precision to all the elements of the Gaussian field.
### --- 2. Simulating from a Dirichlet likelihood --- ####
set.seed(1000)
N <- 100 #number of data
V <- as.data.frame(matrix(runif((4) * N, 0, 1), ncol = 4)) #Covariates
names(V) <- paste0('v', 1:4)
formula <- y ~ 1 + v1 | 1 + v2 | 1 + v3
(names_cat <- formula_list(formula))
intercepts <-
x <- c(-1.5, 1, #Cat 1
-2, 2.3, #Cat 2
0 , -1.9) #Cat 3
mus <- exp(x) / sum(exp(x))
C <- length(names_cat)
data_stack_construct <-
data_stack_dirich(y = as.vector(rep(NA, N * C)),
covariates = names_cat,
data = V,
d = C,
n = N)
A_construct <- data_stack_construct
A_construct[1:8, ]
eta <- A_construct %*% x
alpha <- exp(eta)
alpha <- matrix(alpha,
ncol = C,
byrow = TRUE)
y_o <- rdirichlet(N, alpha)
colnames(y_o) <- paste0("y", 1:C)
head(y_o)Fitting the model
The next step is to call the dirinlareg function in order to fit a model to the data. We just need to specify the formula, the response variable, the covariates and the precision for the Gaussian prior distribution of the parameters.
### --- 3. Fitting the model --- ####
y <- y_o
model.inla <- dirinlareg(
formula = y ~ 1 + v1 | 1 + v2 | 1 + v3,
y = y,
data.cov = V,
prec = 0.0001,
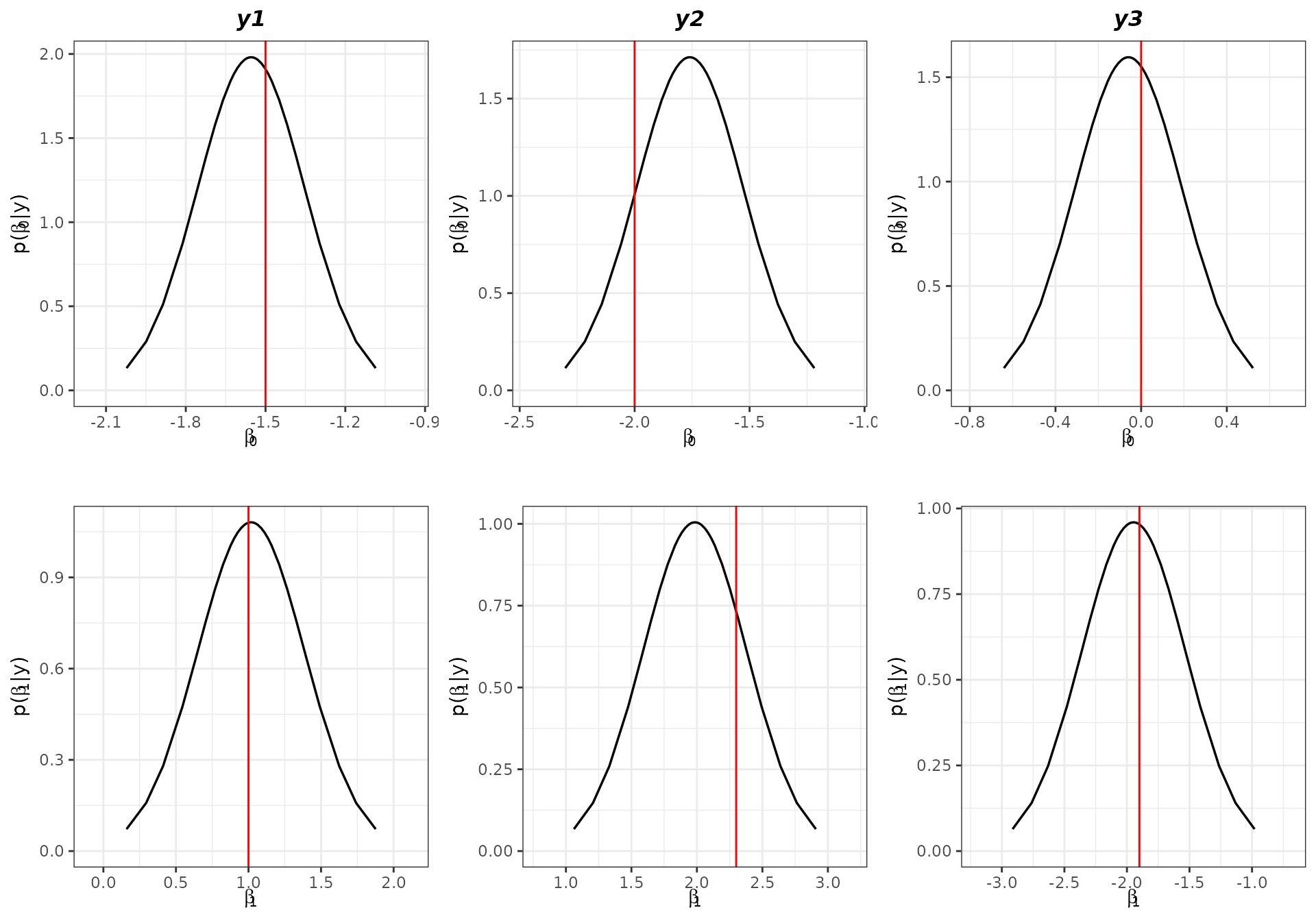
verbose = TRUE)To collect information about the fitted values and marginal posterior distributions of the parameters, we can use the methods summary and plot directly to the dirinlaregmodel object generated.
Summary
summary(model.inla)
#>
#> Call:
#> dirinlareg(formula = y ~ 1 + v1 | 1 + v2 | 1 + v3, y = y, data.cov = V,
#> prec = 1e-04, verbose = TRUE)
#>
#>
#> ---- FIXED EFFECTS ----
#> =======================================================================
#> y1
#> -----------------------------------------------------------------------
#> mean sd 0.025quant 0.5quant 0.975quant mode
#> intercept -1.554 0.2014 -1.9490 -1.554 -1.159 -1.554
#> v1 1.018 0.3690 0.2948 1.018 1.741 1.018
#> =======================================================================
#> y2
#> -----------------------------------------------------------------------
#> mean sd 0.025quant 0.5quant 0.975quant mode
#> intercept -1.760 0.2329 -2.217 -1.760 -1.304 -1.760
#> v2 1.985 0.3971 1.206 1.985 2.763 1.985
#> =======================================================================
#> y3
#> -----------------------------------------------------------------------
#> mean sd 0.025quant 0.5quant 0.975quant mode
#> intercept -0.05927 0.2500 -0.5493 -0.05927 0.4307 -0.05927
#> v3 -1.94703 0.4158 -2.7619 -1.94703 -1.1322 -1.94703
#> =======================================================================
#>
#> ---- HYPERPARAMETERS ----
#>
#> No hyperparameters in the model
#> =======================================================================
#> DIC = 1821.0264 , WAIC = 1711.0295 , LCPO = 913.8332
#> Number of observations: 100
#> Number of Categories: 3Predictions
The package provides a method predict to compute posterior predictive distributions for new individuals. To show how this function works, we will predict for a value of v1 = 0.2, v2 = 0.5, and v3 = -0.1:
### --- 5. Predicting for v1 = 0.25, v2 = 0.5, v3 = 0.5, v4 = 0.1 --- ####
model.prediction <-
predict(model.inla,
data.pred.cov = data.frame(v1 = 0.2 ,
v2 = 0.5,
v3 = -0.1))
#>
#>
#> ---------------------- Predicting -----------------
#>
#>
model.prediction$summary_predictive_means
#> $y1
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> [1,] 0.05081676 0.1148319 0.1372829 0.1405741 0.1619588 0.3298339
#>
#> $y2
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> [1,] 0.07885047 0.203052 0.2470197 0.2529631 0.2966277 0.5925871
#>
#> $y3
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> [1,] 0.3085924 0.5549312 0.6094127 0.6064628 0.6599187 0.8312106
### --- 6. We can also predict directly --- ####
model.inla <- dirinlareg(
formula = y ~ 1 + v1 | 1 + v2 | 1 + v3,
y = y,
data.cov = V,
prec = 0.0001,
verbose = FALSE,
prediction = TRUE,
data.pred.cov = data.frame(v1 = 0.2 ,
v2 = 0.5,
v3 = -0.1))
#>
#>
#> ---------------------- Looking for the mode -----------------
#>
#>
#> ---------------------- INLA call -----------------
#>
#> ---------------------- Obtaining linear predictor -----------------
#>
#>
#> ---------------------- Predicting -----------------
#>
#>
model.prediction$summary_predictive_means
#> $y1
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> [1,] 0.05081676 0.1148319 0.1372829 0.1405741 0.1619588 0.3298339
#>
#> $y2
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> [1,] 0.07885047 0.203052 0.2470197 0.2529631 0.2966277 0.5925871
#>
#> $y3
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> [1,] 0.3085924 0.5549312 0.6094127 0.6064628 0.6599187 0.8312106